
Sewage-associated circular DNA virus-24
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.03
Get precalculated fractions of proteins
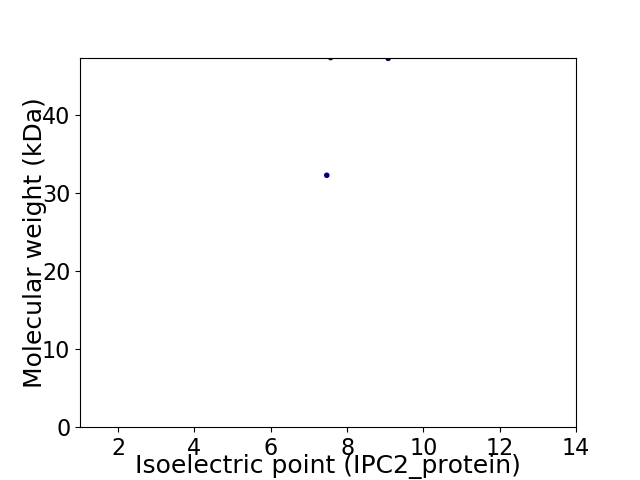
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UI46|A0A0B4UI46_9VIRU Putative capsid protein OS=Sewage-associated circular DNA virus-24 OX=1592091 PE=4 SV=1
MM1 pKa = 7.37SRR3 pKa = 11.84SQHH6 pKa = 6.28LYY8 pKa = 8.63WIGTISIQSFPTWNPEE24 pKa = 3.85EE25 pKa = 4.73YY26 pKa = 10.37FSQTDD31 pKa = 3.3DD32 pKa = 3.43LQYY35 pKa = 10.81IRR37 pKa = 11.84GQKK40 pKa = 10.06EE41 pKa = 3.54EE42 pKa = 4.63GEE44 pKa = 4.63GGFQHH49 pKa = 6.14WQVLMVFSKK58 pKa = 10.57KK59 pKa = 9.9KK60 pKa = 7.89RR61 pKa = 11.84LSWITSRR68 pKa = 11.84TPGHH72 pKa = 6.59WEE74 pKa = 3.55PTRR77 pKa = 11.84SVAADD82 pKa = 3.12QYY84 pKa = 9.42VWKK87 pKa = 10.34EE88 pKa = 3.62EE89 pKa = 3.89TAIPNSRR96 pKa = 11.84FEE98 pKa = 4.01FGEE101 pKa = 3.54RR102 pKa = 11.84RR103 pKa = 11.84IRR105 pKa = 11.84RR106 pKa = 11.84NSKK109 pKa = 8.41TDD111 pKa = 3.1WAIVLQAARR120 pKa = 11.84EE121 pKa = 3.88RR122 pKa = 11.84RR123 pKa = 11.84LGDD126 pKa = 3.18IPEE129 pKa = 5.29DD130 pKa = 3.22IVVRR134 pKa = 11.84YY135 pKa = 8.61YY136 pKa = 11.7GNIIRR141 pKa = 11.84IGSDD145 pKa = 2.54NLRR148 pKa = 11.84PIEE151 pKa = 3.9QVRR154 pKa = 11.84TCKK157 pKa = 10.57VFWGLTGTGKK167 pKa = 9.99SRR169 pKa = 11.84RR170 pKa = 11.84AFEE173 pKa = 4.05EE174 pKa = 4.02AGMEE178 pKa = 4.33CYY180 pKa = 10.33VKK182 pKa = 10.73NPRR185 pKa = 11.84TKK187 pKa = 9.31WWDD190 pKa = 3.39GYY192 pKa = 11.16AGQKK196 pKa = 10.03NVVIDD201 pKa = 3.85EE202 pKa = 4.21FRR204 pKa = 11.84GGIDD208 pKa = 3.41VAYY211 pKa = 10.7LLLWLDD217 pKa = 3.75RR218 pKa = 11.84YY219 pKa = 10.53KK220 pKa = 11.24VIVEE224 pKa = 4.19KK225 pKa = 10.93KK226 pKa = 10.39GGALCLEE233 pKa = 4.85ALQFWITSNISPKK246 pKa = 10.36DD247 pKa = 3.47WYY249 pKa = 10.23PEE251 pKa = 4.25LDD253 pKa = 4.78DD254 pKa = 3.99EE255 pKa = 4.8TQGALRR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84LTEE266 pKa = 3.82VVHH269 pKa = 6.58FDD271 pKa = 3.46VAFEE275 pKa = 4.0
MM1 pKa = 7.37SRR3 pKa = 11.84SQHH6 pKa = 6.28LYY8 pKa = 8.63WIGTISIQSFPTWNPEE24 pKa = 3.85EE25 pKa = 4.73YY26 pKa = 10.37FSQTDD31 pKa = 3.3DD32 pKa = 3.43LQYY35 pKa = 10.81IRR37 pKa = 11.84GQKK40 pKa = 10.06EE41 pKa = 3.54EE42 pKa = 4.63GEE44 pKa = 4.63GGFQHH49 pKa = 6.14WQVLMVFSKK58 pKa = 10.57KK59 pKa = 9.9KK60 pKa = 7.89RR61 pKa = 11.84LSWITSRR68 pKa = 11.84TPGHH72 pKa = 6.59WEE74 pKa = 3.55PTRR77 pKa = 11.84SVAADD82 pKa = 3.12QYY84 pKa = 9.42VWKK87 pKa = 10.34EE88 pKa = 3.62EE89 pKa = 3.89TAIPNSRR96 pKa = 11.84FEE98 pKa = 4.01FGEE101 pKa = 3.54RR102 pKa = 11.84RR103 pKa = 11.84IRR105 pKa = 11.84RR106 pKa = 11.84NSKK109 pKa = 8.41TDD111 pKa = 3.1WAIVLQAARR120 pKa = 11.84EE121 pKa = 3.88RR122 pKa = 11.84RR123 pKa = 11.84LGDD126 pKa = 3.18IPEE129 pKa = 5.29DD130 pKa = 3.22IVVRR134 pKa = 11.84YY135 pKa = 8.61YY136 pKa = 11.7GNIIRR141 pKa = 11.84IGSDD145 pKa = 2.54NLRR148 pKa = 11.84PIEE151 pKa = 3.9QVRR154 pKa = 11.84TCKK157 pKa = 10.57VFWGLTGTGKK167 pKa = 9.99SRR169 pKa = 11.84RR170 pKa = 11.84AFEE173 pKa = 4.05EE174 pKa = 4.02AGMEE178 pKa = 4.33CYY180 pKa = 10.33VKK182 pKa = 10.73NPRR185 pKa = 11.84TKK187 pKa = 9.31WWDD190 pKa = 3.39GYY192 pKa = 11.16AGQKK196 pKa = 10.03NVVIDD201 pKa = 3.85EE202 pKa = 4.21FRR204 pKa = 11.84GGIDD208 pKa = 3.41VAYY211 pKa = 10.7LLLWLDD217 pKa = 3.75RR218 pKa = 11.84YY219 pKa = 10.53KK220 pKa = 11.24VIVEE224 pKa = 4.19KK225 pKa = 10.93KK226 pKa = 10.39GGALCLEE233 pKa = 4.85ALQFWITSNISPKK246 pKa = 10.36DD247 pKa = 3.47WYY249 pKa = 10.23PEE251 pKa = 4.25LDD253 pKa = 4.78DD254 pKa = 3.99EE255 pKa = 4.8TQGALRR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84LTEE266 pKa = 3.82VVHH269 pKa = 6.58FDD271 pKa = 3.46VAFEE275 pKa = 4.0
Molecular weight: 32.31 kDa
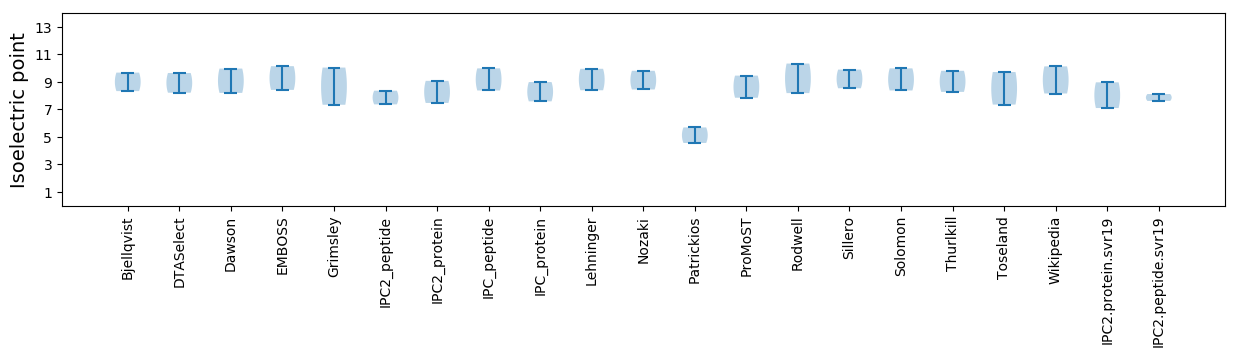
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UI46|A0A0B4UI46_9VIRU Putative capsid protein OS=Sewage-associated circular DNA virus-24 OX=1592091 PE=4 SV=1
MM1 pKa = 7.97GYY3 pKa = 10.29RR4 pKa = 11.84SFVRR8 pKa = 11.84KK9 pKa = 9.1GARR12 pKa = 11.84GLKK15 pKa = 10.0RR16 pKa = 11.84GASRR20 pKa = 11.84YY21 pKa = 9.55RR22 pKa = 11.84SYY24 pKa = 11.45QSGARR29 pKa = 11.84THH31 pKa = 5.74KK32 pKa = 9.24TDD34 pKa = 4.04RR35 pKa = 11.84NSDD38 pKa = 3.38HH39 pKa = 7.02PAVKK43 pKa = 10.23GFRR46 pKa = 11.84TIAGMSRR53 pKa = 11.84KK54 pKa = 9.41ARR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84EE59 pKa = 3.68EE60 pKa = 3.7LGKK63 pKa = 10.66RR64 pKa = 11.84KK65 pKa = 9.49AHH67 pKa = 6.34LSKK70 pKa = 10.8AGQVKK75 pKa = 8.19NTTEE79 pKa = 4.06GVEE82 pKa = 4.38GAVHH86 pKa = 7.34NDD88 pKa = 3.59LQCLNVNFALGHH100 pKa = 5.97AKK102 pKa = 9.81PPKK105 pKa = 10.57GLGKK109 pKa = 9.49WNYY112 pKa = 7.3KK113 pKa = 9.94HH114 pKa = 6.81EE115 pKa = 4.49YY116 pKa = 8.98GVCLFADD123 pKa = 4.69LPMKK127 pKa = 10.48TGVQGQASLWTTCGVQNFIGAPTANNNSFEE157 pKa = 4.21WSGNPFDD164 pKa = 5.49LNPYY168 pKa = 9.67QYY170 pKa = 7.84TTGGVIQAASTDD182 pKa = 3.72KK183 pKa = 11.37NHH185 pKa = 6.44IQTDD189 pKa = 3.93VVRR192 pKa = 11.84MNSIKK197 pKa = 11.11GEE199 pKa = 4.18MNFTNLTVEE208 pKa = 4.25TNRR211 pKa = 11.84LDD213 pKa = 3.85VYY215 pKa = 10.85FMICSSNHH223 pKa = 5.65NLTVTQAWDD232 pKa = 3.78TILTSEE238 pKa = 5.32ALNQSAGSAATIVNGAVVAGYY259 pKa = 9.88VNPSSPFSGGSINSYY274 pKa = 10.41GEE276 pKa = 4.21EE277 pKa = 3.68FTKK280 pKa = 10.74YY281 pKa = 8.69YY282 pKa = 10.78RR283 pKa = 11.84SMRR286 pKa = 11.84KK287 pKa = 8.03FWKK290 pKa = 9.71IKK292 pKa = 10.42KK293 pKa = 9.99KK294 pKa = 9.62LTLCLSSGQTKK305 pKa = 9.91RR306 pKa = 11.84LRR308 pKa = 11.84YY309 pKa = 9.23NIPINKK315 pKa = 8.73VLHH318 pKa = 6.04KK319 pKa = 10.37EE320 pKa = 3.75VLIAMNSTGIQKK332 pKa = 10.12VKK334 pKa = 10.31GLTMEE339 pKa = 4.47CFVISRR345 pKa = 11.84GSPVKK350 pKa = 10.97VFDD353 pKa = 3.74TAGTTMSPALLEE365 pKa = 4.12YY366 pKa = 10.85GFTNLAQYY374 pKa = 9.88QFQACKK380 pKa = 10.11GAAEE384 pKa = 3.81RR385 pKa = 11.84LSFNRR390 pKa = 11.84QFEE393 pKa = 4.53GFVQEE398 pKa = 4.22ATAGASEE405 pKa = 4.34AFLNGADD412 pKa = 4.29IIDD415 pKa = 3.97SAVIATDD422 pKa = 3.16VLTTVAAAMVV432 pKa = 3.61
MM1 pKa = 7.97GYY3 pKa = 10.29RR4 pKa = 11.84SFVRR8 pKa = 11.84KK9 pKa = 9.1GARR12 pKa = 11.84GLKK15 pKa = 10.0RR16 pKa = 11.84GASRR20 pKa = 11.84YY21 pKa = 9.55RR22 pKa = 11.84SYY24 pKa = 11.45QSGARR29 pKa = 11.84THH31 pKa = 5.74KK32 pKa = 9.24TDD34 pKa = 4.04RR35 pKa = 11.84NSDD38 pKa = 3.38HH39 pKa = 7.02PAVKK43 pKa = 10.23GFRR46 pKa = 11.84TIAGMSRR53 pKa = 11.84KK54 pKa = 9.41ARR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84EE59 pKa = 3.68EE60 pKa = 3.7LGKK63 pKa = 10.66RR64 pKa = 11.84KK65 pKa = 9.49AHH67 pKa = 6.34LSKK70 pKa = 10.8AGQVKK75 pKa = 8.19NTTEE79 pKa = 4.06GVEE82 pKa = 4.38GAVHH86 pKa = 7.34NDD88 pKa = 3.59LQCLNVNFALGHH100 pKa = 5.97AKK102 pKa = 9.81PPKK105 pKa = 10.57GLGKK109 pKa = 9.49WNYY112 pKa = 7.3KK113 pKa = 9.94HH114 pKa = 6.81EE115 pKa = 4.49YY116 pKa = 8.98GVCLFADD123 pKa = 4.69LPMKK127 pKa = 10.48TGVQGQASLWTTCGVQNFIGAPTANNNSFEE157 pKa = 4.21WSGNPFDD164 pKa = 5.49LNPYY168 pKa = 9.67QYY170 pKa = 7.84TTGGVIQAASTDD182 pKa = 3.72KK183 pKa = 11.37NHH185 pKa = 6.44IQTDD189 pKa = 3.93VVRR192 pKa = 11.84MNSIKK197 pKa = 11.11GEE199 pKa = 4.18MNFTNLTVEE208 pKa = 4.25TNRR211 pKa = 11.84LDD213 pKa = 3.85VYY215 pKa = 10.85FMICSSNHH223 pKa = 5.65NLTVTQAWDD232 pKa = 3.78TILTSEE238 pKa = 5.32ALNQSAGSAATIVNGAVVAGYY259 pKa = 9.88VNPSSPFSGGSINSYY274 pKa = 10.41GEE276 pKa = 4.21EE277 pKa = 3.68FTKK280 pKa = 10.74YY281 pKa = 8.69YY282 pKa = 10.78RR283 pKa = 11.84SMRR286 pKa = 11.84KK287 pKa = 8.03FWKK290 pKa = 9.71IKK292 pKa = 10.42KK293 pKa = 9.99KK294 pKa = 9.62LTLCLSSGQTKK305 pKa = 9.91RR306 pKa = 11.84LRR308 pKa = 11.84YY309 pKa = 9.23NIPINKK315 pKa = 8.73VLHH318 pKa = 6.04KK319 pKa = 10.37EE320 pKa = 3.75VLIAMNSTGIQKK332 pKa = 10.12VKK334 pKa = 10.31GLTMEE339 pKa = 4.47CFVISRR345 pKa = 11.84GSPVKK350 pKa = 10.97VFDD353 pKa = 3.74TAGTTMSPALLEE365 pKa = 4.12YY366 pKa = 10.85GFTNLAQYY374 pKa = 9.88QFQACKK380 pKa = 10.11GAAEE384 pKa = 3.81RR385 pKa = 11.84LSFNRR390 pKa = 11.84QFEE393 pKa = 4.53GFVQEE398 pKa = 4.22ATAGASEE405 pKa = 4.34AFLNGADD412 pKa = 4.29IIDD415 pKa = 3.97SAVIATDD422 pKa = 3.16VLTTVAAAMVV432 pKa = 3.61
Molecular weight: 47.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
707 |
275 |
432 |
353.5 |
39.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.779 ± 1.732 | 1.414 ± 0.208 |
3.96 ± 0.962 | 5.799 ± 1.652 |
4.526 ± 0.105 | 8.487 ± 0.548 |
1.839 ± 0.247 | 5.233 ± 1.079 |
6.223 ± 0.495 | 6.648 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.98 ± 0.573 | 5.233 ± 1.497 |
3.112 ± 0.338 | 4.385 ± 0.221 |
6.789 ± 1.483 | 6.648 ± 0.769 |
6.931 ± 0.951 | 6.789 ± 0.157 |
2.546 ± 1.405 | 3.678 ± 0.208 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
