
Parabacteroides sp. CAG:409
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; environmental samples
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
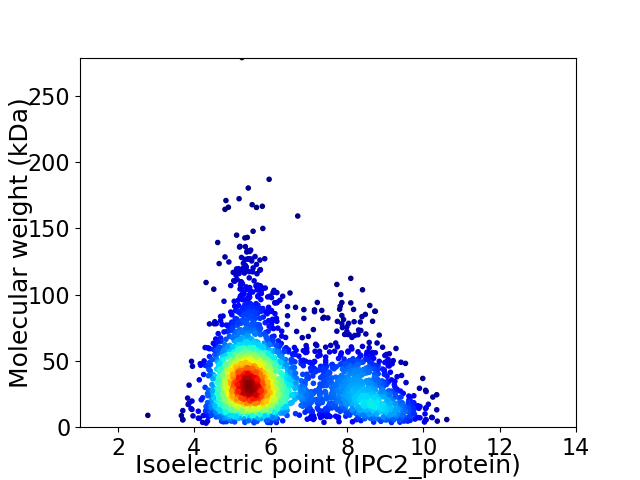
Virtual 2D-PAGE plot for 2823 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7JMQ7|R7JMQ7_9BACT TPR_REGION domain-containing protein OS=Parabacteroides sp. CAG:409 OX=1262913 GN=BN646_02786 PE=4 SV=1
MM1 pKa = 7.04KK2 pKa = 10.05TSNLKK7 pKa = 9.71FLSMAAISAMIAFTSCDD24 pKa = 3.56EE25 pKa = 5.68DD26 pKa = 5.85DD27 pKa = 4.31PTDD30 pKa = 3.68NPNTDD35 pKa = 2.96GTEE38 pKa = 4.13NPDD41 pKa = 3.58GEE43 pKa = 4.49ATGYY47 pKa = 10.47SSEE50 pKa = 4.34LFQAEE55 pKa = 4.39ANHH58 pKa = 5.44NTINFDD64 pKa = 3.89FSNTFSNLFVGTIDD78 pKa = 4.6GGADD82 pKa = 3.29MPSVNSFFTAAAYY95 pKa = 9.83QGAVSADD102 pKa = 3.84NNWTAGWTLTTEE114 pKa = 3.94EE115 pKa = 4.8GGDD118 pKa = 3.33NKK120 pKa = 10.79ADD122 pKa = 3.63QEE124 pKa = 4.32AVEE127 pKa = 4.5TLTGTLSEE135 pKa = 4.86DD136 pKa = 3.28KK137 pKa = 10.63TLEE140 pKa = 4.06ANKK143 pKa = 10.59KK144 pKa = 9.34YY145 pKa = 9.58MLSGDD150 pKa = 4.17YY151 pKa = 10.2IVEE154 pKa = 4.23TGATLTIPEE163 pKa = 4.49GVTIVAEE170 pKa = 4.21ADD172 pKa = 3.59DD173 pKa = 5.37DD174 pKa = 4.07KK175 pKa = 11.82VDD177 pKa = 4.41YY178 pKa = 11.0ILIQQGAKK186 pKa = 9.55IVAEE190 pKa = 4.49GTAEE194 pKa = 3.95NPIVMTATKK203 pKa = 10.15KK204 pKa = 10.31EE205 pKa = 3.68AGAWGGIHH213 pKa = 6.73ICGKK217 pKa = 9.29AHH219 pKa = 6.51SNVEE223 pKa = 3.9GGTGKK228 pKa = 10.21SEE230 pKa = 4.39IGDD233 pKa = 3.51ATYY236 pKa = 10.97GGNDD240 pKa = 3.52DD241 pKa = 4.6ADD243 pKa = 3.46NSGVLKK249 pKa = 9.86YY250 pKa = 10.89LRR252 pKa = 11.84IEE254 pKa = 3.93YY255 pKa = 10.08SGYY258 pKa = 11.01AFDD261 pKa = 6.0SEE263 pKa = 4.99HH264 pKa = 6.46EE265 pKa = 4.31ANGFTFYY272 pKa = 11.24GVGNGTTIEE281 pKa = 4.01YY282 pKa = 9.15CQAYY286 pKa = 9.13KK287 pKa = 10.93GSDD290 pKa = 3.2DD291 pKa = 3.48GFEE294 pKa = 4.15FFGGSVNVKK303 pKa = 9.97NVIAVSCSDD312 pKa = 5.48DD313 pKa = 3.8SFDD316 pKa = 3.8WTEE319 pKa = 3.82GWKK322 pKa = 11.09GNGQFMIAYY331 pKa = 9.2QEE333 pKa = 4.32AEE335 pKa = 3.93EE336 pKa = 4.28SLGYY340 pKa = 10.84DD341 pKa = 3.91CDD343 pKa = 5.24CLMEE347 pKa = 5.18CDD349 pKa = 3.92NNGDD353 pKa = 3.66NFAATPVAHH362 pKa = 6.59PTLANLTLIGNGGSKK377 pKa = 10.06QGIRR381 pKa = 11.84LRR383 pKa = 11.84AGTQASIYY391 pKa = 9.12NALVAGKK398 pKa = 9.68GMCLTTEE405 pKa = 3.95TAEE408 pKa = 4.26TEE410 pKa = 4.13DD411 pKa = 3.92ALVEE415 pKa = 4.31GTSVLNYY422 pKa = 8.22ITLATDD428 pKa = 3.9ITCKK432 pKa = 10.34GAEE435 pKa = 3.89
MM1 pKa = 7.04KK2 pKa = 10.05TSNLKK7 pKa = 9.71FLSMAAISAMIAFTSCDD24 pKa = 3.56EE25 pKa = 5.68DD26 pKa = 5.85DD27 pKa = 4.31PTDD30 pKa = 3.68NPNTDD35 pKa = 2.96GTEE38 pKa = 4.13NPDD41 pKa = 3.58GEE43 pKa = 4.49ATGYY47 pKa = 10.47SSEE50 pKa = 4.34LFQAEE55 pKa = 4.39ANHH58 pKa = 5.44NTINFDD64 pKa = 3.89FSNTFSNLFVGTIDD78 pKa = 4.6GGADD82 pKa = 3.29MPSVNSFFTAAAYY95 pKa = 9.83QGAVSADD102 pKa = 3.84NNWTAGWTLTTEE114 pKa = 3.94EE115 pKa = 4.8GGDD118 pKa = 3.33NKK120 pKa = 10.79ADD122 pKa = 3.63QEE124 pKa = 4.32AVEE127 pKa = 4.5TLTGTLSEE135 pKa = 4.86DD136 pKa = 3.28KK137 pKa = 10.63TLEE140 pKa = 4.06ANKK143 pKa = 10.59KK144 pKa = 9.34YY145 pKa = 9.58MLSGDD150 pKa = 4.17YY151 pKa = 10.2IVEE154 pKa = 4.23TGATLTIPEE163 pKa = 4.49GVTIVAEE170 pKa = 4.21ADD172 pKa = 3.59DD173 pKa = 5.37DD174 pKa = 4.07KK175 pKa = 11.82VDD177 pKa = 4.41YY178 pKa = 11.0ILIQQGAKK186 pKa = 9.55IVAEE190 pKa = 4.49GTAEE194 pKa = 3.95NPIVMTATKK203 pKa = 10.15KK204 pKa = 10.31EE205 pKa = 3.68AGAWGGIHH213 pKa = 6.73ICGKK217 pKa = 9.29AHH219 pKa = 6.51SNVEE223 pKa = 3.9GGTGKK228 pKa = 10.21SEE230 pKa = 4.39IGDD233 pKa = 3.51ATYY236 pKa = 10.97GGNDD240 pKa = 3.52DD241 pKa = 4.6ADD243 pKa = 3.46NSGVLKK249 pKa = 9.86YY250 pKa = 10.89LRR252 pKa = 11.84IEE254 pKa = 3.93YY255 pKa = 10.08SGYY258 pKa = 11.01AFDD261 pKa = 6.0SEE263 pKa = 4.99HH264 pKa = 6.46EE265 pKa = 4.31ANGFTFYY272 pKa = 11.24GVGNGTTIEE281 pKa = 4.01YY282 pKa = 9.15CQAYY286 pKa = 9.13KK287 pKa = 10.93GSDD290 pKa = 3.2DD291 pKa = 3.48GFEE294 pKa = 4.15FFGGSVNVKK303 pKa = 9.97NVIAVSCSDD312 pKa = 5.48DD313 pKa = 3.8SFDD316 pKa = 3.8WTEE319 pKa = 3.82GWKK322 pKa = 11.09GNGQFMIAYY331 pKa = 9.2QEE333 pKa = 4.32AEE335 pKa = 3.93EE336 pKa = 4.28SLGYY340 pKa = 10.84DD341 pKa = 3.91CDD343 pKa = 5.24CLMEE347 pKa = 5.18CDD349 pKa = 3.92NNGDD353 pKa = 3.66NFAATPVAHH362 pKa = 6.59PTLANLTLIGNGGSKK377 pKa = 10.06QGIRR381 pKa = 11.84LRR383 pKa = 11.84AGTQASIYY391 pKa = 9.12NALVAGKK398 pKa = 9.68GMCLTTEE405 pKa = 3.95TAEE408 pKa = 4.26TEE410 pKa = 4.13DD411 pKa = 3.92ALVEE415 pKa = 4.31GTSVLNYY422 pKa = 8.22ITLATDD428 pKa = 3.9ITCKK432 pKa = 10.34GAEE435 pKa = 3.89
Molecular weight: 45.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7JCG2|R7JCG2_9BACT DUF5672 domain-containing protein OS=Parabacteroides sp. CAG:409 OX=1262913 GN=BN646_01290 PE=4 SV=1
MM1 pKa = 7.25TSAIFSPAATCCPGVTFTKK20 pKa = 10.34LSCPSIGEE28 pKa = 4.18RR29 pKa = 11.84TTKK32 pKa = 10.65SSIRR36 pKa = 11.84CRR38 pKa = 11.84LRR40 pKa = 11.84INFFSSRR47 pKa = 11.84SRR49 pKa = 11.84SFCNNSFRR57 pKa = 11.84ITVMEE62 pKa = 4.03EE63 pKa = 3.91SLWRR67 pKa = 11.84LFNFRR72 pKa = 11.84RR73 pKa = 11.84VDD75 pKa = 3.6SVLYY79 pKa = 9.93RR80 pKa = 11.84YY81 pKa = 10.17SSSDD85 pKa = 3.17TSSWALLRR93 pKa = 11.84IPIAYY98 pKa = 10.2SSFSSEE104 pKa = 3.61KK105 pKa = 10.49LFRR108 pKa = 11.84ILFNSYY114 pKa = 9.85SASNVCCFKK123 pKa = 10.96VRR125 pKa = 11.84LFCSISRR132 pKa = 11.84AFSS135 pKa = 3.18
MM1 pKa = 7.25TSAIFSPAATCCPGVTFTKK20 pKa = 10.34LSCPSIGEE28 pKa = 4.18RR29 pKa = 11.84TTKK32 pKa = 10.65SSIRR36 pKa = 11.84CRR38 pKa = 11.84LRR40 pKa = 11.84INFFSSRR47 pKa = 11.84SRR49 pKa = 11.84SFCNNSFRR57 pKa = 11.84ITVMEE62 pKa = 4.03EE63 pKa = 3.91SLWRR67 pKa = 11.84LFNFRR72 pKa = 11.84RR73 pKa = 11.84VDD75 pKa = 3.6SVLYY79 pKa = 9.93RR80 pKa = 11.84YY81 pKa = 10.17SSSDD85 pKa = 3.17TSSWALLRR93 pKa = 11.84IPIAYY98 pKa = 10.2SSFSSEE104 pKa = 3.61KK105 pKa = 10.49LFRR108 pKa = 11.84ILFNSYY114 pKa = 9.85SASNVCCFKK123 pKa = 10.96VRR125 pKa = 11.84LFCSISRR132 pKa = 11.84AFSS135 pKa = 3.18
Molecular weight: 15.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
967119 |
30 |
2471 |
342.6 |
38.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.095 ± 0.041 | 1.238 ± 0.016 |
5.483 ± 0.029 | 6.71 ± 0.045 |
4.585 ± 0.034 | 6.773 ± 0.045 |
1.955 ± 0.02 | 6.843 ± 0.044 |
6.421 ± 0.046 | 9.307 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.72 ± 0.021 | 4.783 ± 0.032 |
3.907 ± 0.028 | 3.963 ± 0.029 |
4.504 ± 0.03 | 6.035 ± 0.035 |
5.383 ± 0.031 | 6.509 ± 0.034 |
1.337 ± 0.018 | 4.429 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
