
Tortoise microvirus 14
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.17
Get precalculated fractions of proteins
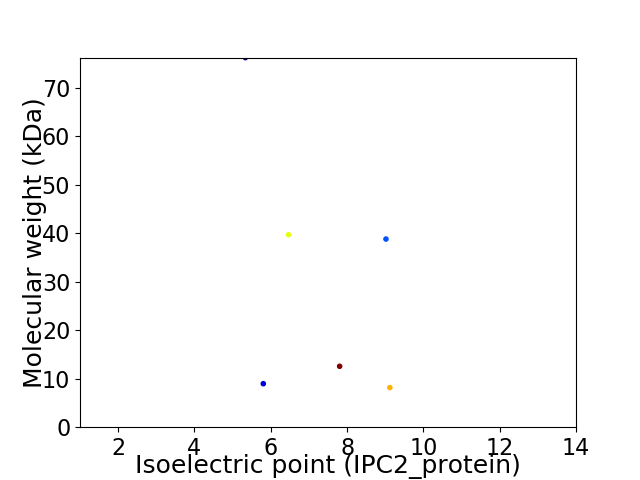
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
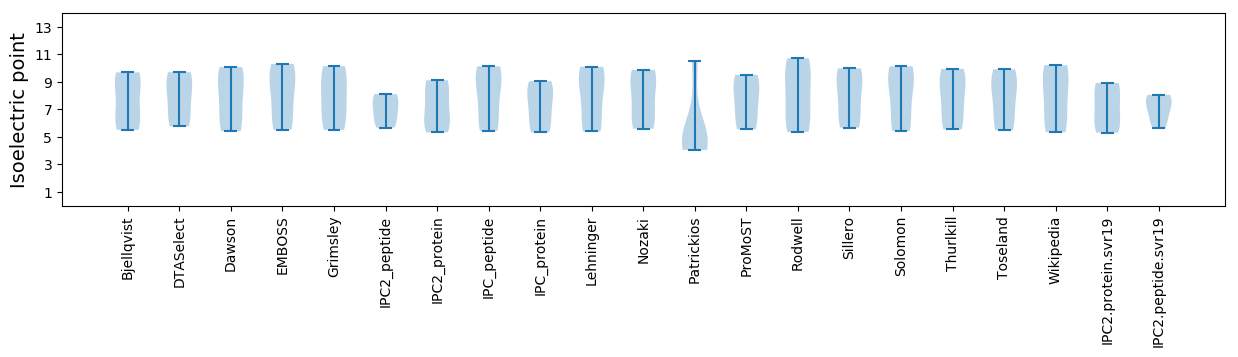
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W653|A0A4P8W653_9VIRU Uncharacterized protein OS=Tortoise microvirus 14 OX=2583115 PE=4 SV=1
MM1 pKa = 7.54SKK3 pKa = 9.0TVNIGGDD10 pKa = 3.26RR11 pKa = 11.84LGSGNKK17 pKa = 9.15MNVQLHH23 pKa = 6.16GYY25 pKa = 9.01SRR27 pKa = 11.84SNHH30 pKa = 6.01DD31 pKa = 3.97LSYY34 pKa = 10.17VWRR37 pKa = 11.84STMSPGTLVPFMVQVALPGDD57 pKa = 3.65SHH59 pKa = 8.64DD60 pKa = 3.91IEE62 pKa = 5.41LNAEE66 pKa = 4.33VMTHH70 pKa = 5.88PTLGPLFGSYY80 pKa = 10.52KK81 pKa = 10.09LQLDD85 pKa = 3.8VFSIPFRR92 pKa = 11.84LYY94 pKa = 11.06NSWLHH99 pKa = 5.48NNRR102 pKa = 11.84LGIGLDD108 pKa = 3.27MSKK111 pKa = 10.93VKK113 pKa = 10.65LPKK116 pKa = 10.47LKK118 pKa = 10.81FNLPYY123 pKa = 10.53INATGSNIPVDD134 pKa = 3.91EE135 pKa = 4.04QQINPSSLLAYY146 pKa = 10.32LGIRR150 pKa = 11.84GGIRR154 pKa = 11.84TNTGQISSPYY164 pKa = 9.67RR165 pKa = 11.84RR166 pKa = 11.84NTKK169 pKa = 8.89MAIPYY174 pKa = 8.91LAYY177 pKa = 8.98WDD179 pKa = 3.77IYY181 pKa = 10.0KK182 pKa = 10.56NYY184 pKa = 9.9YY185 pKa = 9.6SNKK188 pKa = 8.24QEE190 pKa = 4.1EE191 pKa = 4.58IGVFIHH197 pKa = 6.77NSGEE201 pKa = 4.24VPQITQLLQAIRR213 pKa = 11.84SRR215 pKa = 11.84GGTIEE220 pKa = 5.78LIFTSSTLPYY230 pKa = 10.38QGILVPLGSDD240 pKa = 3.23LTIQNLPGSATAEE253 pKa = 3.68MLRR256 pKa = 11.84FNDD259 pKa = 3.44TRR261 pKa = 11.84SGEE264 pKa = 4.1SGTWDD269 pKa = 3.21KK270 pKa = 11.23FFNNYY275 pKa = 6.58TTTLTPSGTRR285 pKa = 11.84VIQFLDD291 pKa = 3.3SKK293 pKa = 10.87YY294 pKa = 10.45EE295 pKa = 3.99NEE297 pKa = 4.32AGTGFEE303 pKa = 5.2GFIDD307 pKa = 4.02QNPQIIPNTAEE318 pKa = 3.67ITLKK322 pKa = 10.47EE323 pKa = 4.17FPLEE327 pKa = 4.56NIDD330 pKa = 4.12NMRR333 pKa = 11.84DD334 pKa = 3.37KK335 pKa = 10.91ILAQQGNIEE344 pKa = 4.26FEE346 pKa = 4.25LMEE349 pKa = 4.66NQPSPYY355 pKa = 10.16GDD357 pKa = 3.78PLLFYY362 pKa = 10.48PDD364 pKa = 3.81TNKK367 pKa = 10.55SNSFFNLEE375 pKa = 3.73GLAVKK380 pKa = 9.15TYY382 pKa = 10.71QSDD385 pKa = 3.92IFNNWINTEE394 pKa = 3.94WLDD397 pKa = 3.92GEE399 pKa = 4.68NGINTITAVDD409 pKa = 3.86VSSGFLNLDD418 pKa = 3.54SLNLAKK424 pKa = 10.25KK425 pKa = 10.31VYY427 pKa = 11.1DD428 pKa = 3.56MLNRR432 pKa = 11.84IAISGGSYY440 pKa = 10.56KK441 pKa = 10.71DD442 pKa = 3.14WLEE445 pKa = 4.18SVWMHH450 pKa = 6.14EE451 pKa = 4.46YY452 pKa = 10.27ISNAEE457 pKa = 4.07SPVYY461 pKa = 10.33EE462 pKa = 4.74GGLSKK467 pKa = 10.68EE468 pKa = 4.16IVFSEE473 pKa = 4.6VVSNSSGIGEE483 pKa = 4.49DD484 pKa = 4.47SSNQPLGTLAGRR496 pKa = 11.84GHH498 pKa = 7.83LSGKK502 pKa = 9.8HH503 pKa = 5.31KK504 pKa = 10.39GGHH507 pKa = 5.68IIIRR511 pKa = 11.84TNEE514 pKa = 3.53PSLIMGIVSITPRR527 pKa = 11.84IDD529 pKa = 3.26YY530 pKa = 10.52SQGNEE535 pKa = 3.52WFTNLDD541 pKa = 3.6TMDD544 pKa = 5.27DD545 pKa = 3.63LHH547 pKa = 8.3KK548 pKa = 10.67PGLDD552 pKa = 3.44GIGFQEE558 pKa = 4.75LVTEE562 pKa = 5.45QFAAWDD568 pKa = 4.34TVWDD572 pKa = 4.03TNEE575 pKa = 3.56DD576 pKa = 3.48RR577 pKa = 11.84VIKK580 pKa = 10.34FSAGKK585 pKa = 8.74QPAWINYY592 pKa = 4.33MTNYY596 pKa = 9.25NRR598 pKa = 11.84VYY600 pKa = 11.34GNFAKK605 pKa = 10.56KK606 pKa = 9.86NNEE609 pKa = 3.38MFMTLTRR616 pKa = 11.84RR617 pKa = 11.84FSYY620 pKa = 10.8NKK622 pKa = 9.5NAANNKK628 pKa = 9.57SAIEE632 pKa = 4.14DD633 pKa = 3.54LTTYY637 pKa = 8.45VQPDD641 pKa = 3.01KK642 pKa = 11.23HH643 pKa = 7.01QDD645 pKa = 2.87TFAITTIDD653 pKa = 3.42AQNFWTQIAVKK664 pKa = 9.89CFARR668 pKa = 11.84RR669 pKa = 11.84KK670 pKa = 8.28MSAKK674 pKa = 10.15IIPNLL679 pKa = 3.51
MM1 pKa = 7.54SKK3 pKa = 9.0TVNIGGDD10 pKa = 3.26RR11 pKa = 11.84LGSGNKK17 pKa = 9.15MNVQLHH23 pKa = 6.16GYY25 pKa = 9.01SRR27 pKa = 11.84SNHH30 pKa = 6.01DD31 pKa = 3.97LSYY34 pKa = 10.17VWRR37 pKa = 11.84STMSPGTLVPFMVQVALPGDD57 pKa = 3.65SHH59 pKa = 8.64DD60 pKa = 3.91IEE62 pKa = 5.41LNAEE66 pKa = 4.33VMTHH70 pKa = 5.88PTLGPLFGSYY80 pKa = 10.52KK81 pKa = 10.09LQLDD85 pKa = 3.8VFSIPFRR92 pKa = 11.84LYY94 pKa = 11.06NSWLHH99 pKa = 5.48NNRR102 pKa = 11.84LGIGLDD108 pKa = 3.27MSKK111 pKa = 10.93VKK113 pKa = 10.65LPKK116 pKa = 10.47LKK118 pKa = 10.81FNLPYY123 pKa = 10.53INATGSNIPVDD134 pKa = 3.91EE135 pKa = 4.04QQINPSSLLAYY146 pKa = 10.32LGIRR150 pKa = 11.84GGIRR154 pKa = 11.84TNTGQISSPYY164 pKa = 9.67RR165 pKa = 11.84RR166 pKa = 11.84NTKK169 pKa = 8.89MAIPYY174 pKa = 8.91LAYY177 pKa = 8.98WDD179 pKa = 3.77IYY181 pKa = 10.0KK182 pKa = 10.56NYY184 pKa = 9.9YY185 pKa = 9.6SNKK188 pKa = 8.24QEE190 pKa = 4.1EE191 pKa = 4.58IGVFIHH197 pKa = 6.77NSGEE201 pKa = 4.24VPQITQLLQAIRR213 pKa = 11.84SRR215 pKa = 11.84GGTIEE220 pKa = 5.78LIFTSSTLPYY230 pKa = 10.38QGILVPLGSDD240 pKa = 3.23LTIQNLPGSATAEE253 pKa = 3.68MLRR256 pKa = 11.84FNDD259 pKa = 3.44TRR261 pKa = 11.84SGEE264 pKa = 4.1SGTWDD269 pKa = 3.21KK270 pKa = 11.23FFNNYY275 pKa = 6.58TTTLTPSGTRR285 pKa = 11.84VIQFLDD291 pKa = 3.3SKK293 pKa = 10.87YY294 pKa = 10.45EE295 pKa = 3.99NEE297 pKa = 4.32AGTGFEE303 pKa = 5.2GFIDD307 pKa = 4.02QNPQIIPNTAEE318 pKa = 3.67ITLKK322 pKa = 10.47EE323 pKa = 4.17FPLEE327 pKa = 4.56NIDD330 pKa = 4.12NMRR333 pKa = 11.84DD334 pKa = 3.37KK335 pKa = 10.91ILAQQGNIEE344 pKa = 4.26FEE346 pKa = 4.25LMEE349 pKa = 4.66NQPSPYY355 pKa = 10.16GDD357 pKa = 3.78PLLFYY362 pKa = 10.48PDD364 pKa = 3.81TNKK367 pKa = 10.55SNSFFNLEE375 pKa = 3.73GLAVKK380 pKa = 9.15TYY382 pKa = 10.71QSDD385 pKa = 3.92IFNNWINTEE394 pKa = 3.94WLDD397 pKa = 3.92GEE399 pKa = 4.68NGINTITAVDD409 pKa = 3.86VSSGFLNLDD418 pKa = 3.54SLNLAKK424 pKa = 10.25KK425 pKa = 10.31VYY427 pKa = 11.1DD428 pKa = 3.56MLNRR432 pKa = 11.84IAISGGSYY440 pKa = 10.56KK441 pKa = 10.71DD442 pKa = 3.14WLEE445 pKa = 4.18SVWMHH450 pKa = 6.14EE451 pKa = 4.46YY452 pKa = 10.27ISNAEE457 pKa = 4.07SPVYY461 pKa = 10.33EE462 pKa = 4.74GGLSKK467 pKa = 10.68EE468 pKa = 4.16IVFSEE473 pKa = 4.6VVSNSSGIGEE483 pKa = 4.49DD484 pKa = 4.47SSNQPLGTLAGRR496 pKa = 11.84GHH498 pKa = 7.83LSGKK502 pKa = 9.8HH503 pKa = 5.31KK504 pKa = 10.39GGHH507 pKa = 5.68IIIRR511 pKa = 11.84TNEE514 pKa = 3.53PSLIMGIVSITPRR527 pKa = 11.84IDD529 pKa = 3.26YY530 pKa = 10.52SQGNEE535 pKa = 3.52WFTNLDD541 pKa = 3.6TMDD544 pKa = 5.27DD545 pKa = 3.63LHH547 pKa = 8.3KK548 pKa = 10.67PGLDD552 pKa = 3.44GIGFQEE558 pKa = 4.75LVTEE562 pKa = 5.45QFAAWDD568 pKa = 4.34TVWDD572 pKa = 4.03TNEE575 pKa = 3.56DD576 pKa = 3.48RR577 pKa = 11.84VIKK580 pKa = 10.34FSAGKK585 pKa = 8.74QPAWINYY592 pKa = 4.33MTNYY596 pKa = 9.25NRR598 pKa = 11.84VYY600 pKa = 11.34GNFAKK605 pKa = 10.56KK606 pKa = 9.86NNEE609 pKa = 3.38MFMTLTRR616 pKa = 11.84RR617 pKa = 11.84FSYY620 pKa = 10.8NKK622 pKa = 9.5NAANNKK628 pKa = 9.57SAIEE632 pKa = 4.14DD633 pKa = 3.54LTTYY637 pKa = 8.45VQPDD641 pKa = 3.01KK642 pKa = 11.23HH643 pKa = 7.01QDD645 pKa = 2.87TFAITTIDD653 pKa = 3.42AQNFWTQIAVKK664 pKa = 9.89CFARR668 pKa = 11.84RR669 pKa = 11.84KK670 pKa = 8.28MSAKK674 pKa = 10.15IIPNLL679 pKa = 3.51
Molecular weight: 76.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVW7|A0A4V1FVW7_9VIRU DNA pilot protein OS=Tortoise microvirus 14 OX=2583115 PE=4 SV=1
MM1 pKa = 7.66LKK3 pKa = 10.54NNSKK7 pKa = 9.83IKK9 pKa = 10.46NMKK12 pKa = 9.67ISINMEE18 pKa = 3.93TFTLQDD24 pKa = 3.68LKK26 pKa = 10.79ILQSILLRR34 pKa = 11.84YY35 pKa = 9.45KK36 pKa = 9.58STYY39 pKa = 9.24NRR41 pKa = 11.84NALKK45 pKa = 10.52NINKK49 pKa = 10.03LIVDD53 pKa = 5.01IINEE57 pKa = 4.13KK58 pKa = 10.5YY59 pKa = 10.68SIPFEE64 pKa = 4.49PDD66 pKa = 2.75PLFF69 pKa = 4.52
MM1 pKa = 7.66LKK3 pKa = 10.54NNSKK7 pKa = 9.83IKK9 pKa = 10.46NMKK12 pKa = 9.67ISINMEE18 pKa = 3.93TFTLQDD24 pKa = 3.68LKK26 pKa = 10.79ILQSILLRR34 pKa = 11.84YY35 pKa = 9.45KK36 pKa = 9.58STYY39 pKa = 9.24NRR41 pKa = 11.84NALKK45 pKa = 10.52NINKK49 pKa = 10.03LIVDD53 pKa = 5.01IINEE57 pKa = 4.13KK58 pKa = 10.5YY59 pKa = 10.68SIPFEE64 pKa = 4.49PDD66 pKa = 2.75PLFF69 pKa = 4.52
Molecular weight: 8.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1616 |
69 |
679 |
269.3 |
30.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.755 ± 1.538 | 0.433 ± 0.292 |
4.517 ± 0.398 | 7.611 ± 1.445 |
3.465 ± 0.746 | 6.869 ± 0.935 |
1.361 ± 0.264 | 8.478 ± 0.623 |
8.168 ± 2.199 | 7.611 ± 0.788 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.723 ± 0.428 | 7.735 ± 0.617 |
3.527 ± 0.677 | 4.703 ± 1.331 |
5.012 ± 0.792 | 6.559 ± 0.773 |
5.941 ± 0.786 | 3.527 ± 0.718 |
1.361 ± 0.283 | 4.641 ± 0.99 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
