
Tropheryma whipplei (strain Twist) (Whipple s bacillus)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Tropherymataceae; Tropheryma; Tropheryma whipplei
Average proteome isoelectric point is 7.34
Get precalculated fractions of proteins
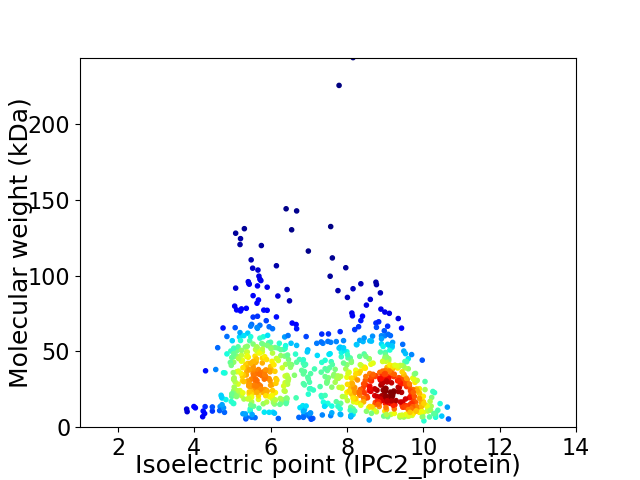
Virtual 2D-PAGE plot for 805 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
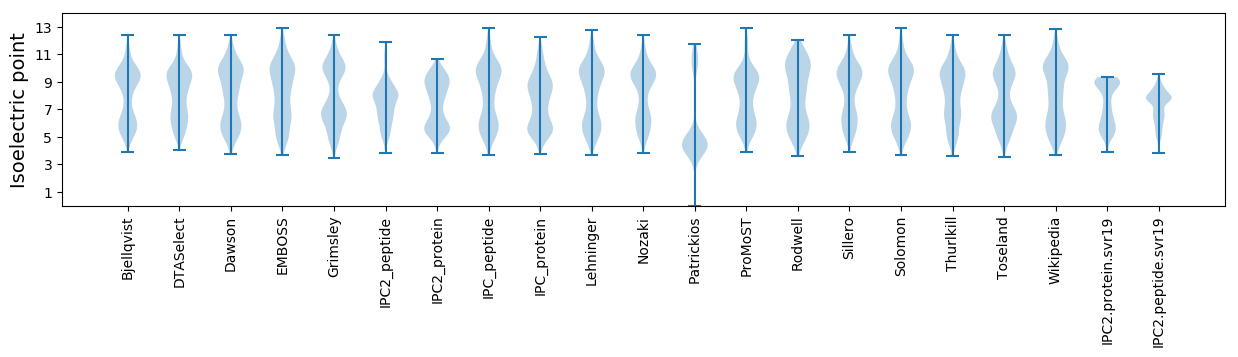
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q83MT9|Q83MT9_TROWT Mg2+ transporter OS=Tropheryma whipplei (strain Twist) OX=203267 GN=mgtE PE=4 SV=1
MM1 pKa = 7.45TYY3 pKa = 10.61VIAFPCVDD11 pKa = 4.09LKK13 pKa = 11.37DD14 pKa = 3.58RR15 pKa = 11.84ACIDD19 pKa = 3.92EE20 pKa = 4.81CPVDD24 pKa = 4.54CIYY27 pKa = 11.16EE28 pKa = 4.34GGRR31 pKa = 11.84SLYY34 pKa = 10.21INPDD38 pKa = 3.04EE39 pKa = 5.27CVDD42 pKa = 4.03CGACEE47 pKa = 3.85PVCPVEE53 pKa = 4.4AIYY56 pKa = 11.22YY57 pKa = 10.15EE58 pKa = 4.88DD59 pKa = 5.24DD60 pKa = 4.53LPEE63 pKa = 4.05EE64 pKa = 3.64WGEE67 pKa = 3.87YY68 pKa = 9.04YY69 pKa = 10.39RR70 pKa = 11.84ANVEE74 pKa = 4.02FFDD77 pKa = 5.29EE78 pKa = 4.48IGSPGGAAKK87 pKa = 10.5LGPVDD92 pKa = 4.41FDD94 pKa = 4.09HH95 pKa = 7.61PIIAQLPKK103 pKa = 10.7SSDD106 pKa = 3.58SVV108 pKa = 3.37
MM1 pKa = 7.45TYY3 pKa = 10.61VIAFPCVDD11 pKa = 4.09LKK13 pKa = 11.37DD14 pKa = 3.58RR15 pKa = 11.84ACIDD19 pKa = 3.92EE20 pKa = 4.81CPVDD24 pKa = 4.54CIYY27 pKa = 11.16EE28 pKa = 4.34GGRR31 pKa = 11.84SLYY34 pKa = 10.21INPDD38 pKa = 3.04EE39 pKa = 5.27CVDD42 pKa = 4.03CGACEE47 pKa = 3.85PVCPVEE53 pKa = 4.4AIYY56 pKa = 11.22YY57 pKa = 10.15EE58 pKa = 4.88DD59 pKa = 5.24DD60 pKa = 4.53LPEE63 pKa = 4.05EE64 pKa = 3.64WGEE67 pKa = 3.87YY68 pKa = 9.04YY69 pKa = 10.39RR70 pKa = 11.84ANVEE74 pKa = 4.02FFDD77 pKa = 5.29EE78 pKa = 4.48IGSPGGAAKK87 pKa = 10.5LGPVDD92 pKa = 4.41FDD94 pKa = 4.09HH95 pKa = 7.61PIIAQLPKK103 pKa = 10.7SSDD106 pKa = 3.58SVV108 pKa = 3.37
Molecular weight: 11.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q83GJ4|Q83GJ4_TROWT Uncharacterized protein OS=Tropheryma whipplei (strain Twist) OX=203267 GN=TWT_277 PE=4 SV=1
MM1 pKa = 7.42AGSRR5 pKa = 11.84FRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 9.18LLQRR14 pKa = 11.84FHH16 pKa = 6.89RR17 pKa = 11.84TNKK20 pKa = 10.11RR21 pKa = 11.84GGLMSRR27 pKa = 11.84ASAKK31 pKa = 9.91SFRR34 pKa = 11.84FAPAQYY40 pKa = 8.96PCVGVTDD47 pKa = 4.5CLGSPKK53 pKa = 9.95YY54 pKa = 10.63VPSGYY59 pKa = 10.17RR60 pKa = 11.84LVEE63 pKa = 3.92ASVSLGSGDD72 pKa = 3.51KK73 pKa = 10.74RR74 pKa = 11.84FFSSRR79 pKa = 11.84EE80 pKa = 3.59LLMCWKK86 pKa = 9.5VHH88 pKa = 6.09RR89 pKa = 11.84LAGLNVRR96 pKa = 11.84VLDD99 pKa = 4.55PGEE102 pKa = 4.08VDD104 pKa = 4.39CLGGSPILRR113 pKa = 11.84DD114 pKa = 3.34KK115 pKa = 11.18EE116 pKa = 4.29RR117 pKa = 11.84ILDD120 pKa = 3.53EE121 pKa = 4.58SGFDD125 pKa = 3.8FARR128 pKa = 11.84PGCIVALRR136 pKa = 11.84SGLRR140 pKa = 11.84RR141 pKa = 11.84VRR143 pKa = 11.84SVMRR147 pKa = 11.84VVYY150 pKa = 10.2AVSDD154 pKa = 3.85VNRR157 pKa = 11.84VALILASIVGHH168 pKa = 5.63GLHH171 pKa = 6.97LEE173 pKa = 3.83EE174 pKa = 5.24GFFIEE179 pKa = 4.2KK180 pKa = 10.55HH181 pKa = 6.06PDD183 pKa = 2.65SSVRR187 pKa = 11.84FTVRR191 pKa = 11.84ILRR194 pKa = 11.84RR195 pKa = 11.84ASCVSWITMNASKK208 pKa = 10.58HH209 pKa = 4.84LVSRR213 pKa = 11.84YY214 pKa = 9.97LRR216 pKa = 11.84VLDD219 pKa = 4.11PTWALL224 pKa = 3.41
MM1 pKa = 7.42AGSRR5 pKa = 11.84FRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 9.18LLQRR14 pKa = 11.84FHH16 pKa = 6.89RR17 pKa = 11.84TNKK20 pKa = 10.11RR21 pKa = 11.84GGLMSRR27 pKa = 11.84ASAKK31 pKa = 9.91SFRR34 pKa = 11.84FAPAQYY40 pKa = 8.96PCVGVTDD47 pKa = 4.5CLGSPKK53 pKa = 9.95YY54 pKa = 10.63VPSGYY59 pKa = 10.17RR60 pKa = 11.84LVEE63 pKa = 3.92ASVSLGSGDD72 pKa = 3.51KK73 pKa = 10.74RR74 pKa = 11.84FFSSRR79 pKa = 11.84EE80 pKa = 3.59LLMCWKK86 pKa = 9.5VHH88 pKa = 6.09RR89 pKa = 11.84LAGLNVRR96 pKa = 11.84VLDD99 pKa = 4.55PGEE102 pKa = 4.08VDD104 pKa = 4.39CLGGSPILRR113 pKa = 11.84DD114 pKa = 3.34KK115 pKa = 11.18EE116 pKa = 4.29RR117 pKa = 11.84ILDD120 pKa = 3.53EE121 pKa = 4.58SGFDD125 pKa = 3.8FARR128 pKa = 11.84PGCIVALRR136 pKa = 11.84SGLRR140 pKa = 11.84RR141 pKa = 11.84VRR143 pKa = 11.84SVMRR147 pKa = 11.84VVYY150 pKa = 10.2AVSDD154 pKa = 3.85VNRR157 pKa = 11.84VALILASIVGHH168 pKa = 5.63GLHH171 pKa = 6.97LEE173 pKa = 3.83EE174 pKa = 5.24GFFIEE179 pKa = 4.2KK180 pKa = 10.55HH181 pKa = 6.06PDD183 pKa = 2.65SSVRR187 pKa = 11.84FTVRR191 pKa = 11.84ILRR194 pKa = 11.84RR195 pKa = 11.84ASCVSWITMNASKK208 pKa = 10.58HH209 pKa = 4.84LVSRR213 pKa = 11.84YY214 pKa = 9.97LRR216 pKa = 11.84VLDD219 pKa = 4.11PTWALL224 pKa = 3.41
Molecular weight: 25.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
263585 |
37 |
2312 |
327.4 |
36.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.332 ± 0.1 | 1.336 ± 0.036 |
5.269 ± 0.076 | 5.022 ± 0.088 |
4.207 ± 0.066 | 7.367 ± 0.073 |
2.081 ± 0.041 | 6.741 ± 0.072 |
4.75 ± 0.062 | 10.263 ± 0.082 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.982 ± 0.036 | 3.578 ± 0.042 |
4.561 ± 0.068 | 3.091 ± 0.042 |
6.291 ± 0.074 | 7.915 ± 0.065 |
5.473 ± 0.131 | 7.945 ± 0.082 |
1.016 ± 0.033 | 2.778 ± 0.051 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
