
Streptomyces phage Jay2Jay
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Samistivirus; Streptomyces virus Jay2Jay
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
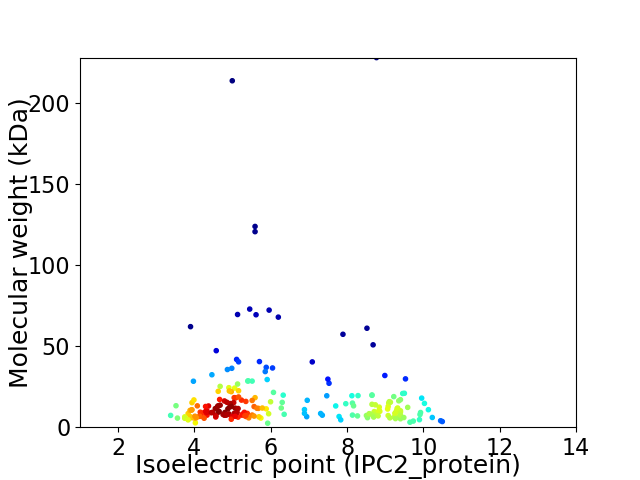
Virtual 2D-PAGE plot for 209 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0RL59|A0A0A0RL59_9CAUD Uncharacterized protein OS=Streptomyces phage Jay2Jay OX=1556290 GN=253 PE=4 SV=1
MM1 pKa = 7.76SYY3 pKa = 10.53QLQVLTDD10 pKa = 3.56APFSYY15 pKa = 9.79WKK17 pKa = 10.51LDD19 pKa = 3.37EE20 pKa = 4.85TGPAFPDD27 pKa = 3.27SAGSMRR33 pKa = 11.84TADD36 pKa = 3.72LVGTITRR43 pKa = 11.84HH44 pKa = 5.3PALVTGSGNALVLDD58 pKa = 4.24NTNHH62 pKa = 7.1LDD64 pKa = 3.3MDD66 pKa = 4.3DD67 pKa = 3.64PVFNKK72 pKa = 10.34GYY74 pKa = 8.92EE75 pKa = 3.89LRR77 pKa = 11.84QFSLEE82 pKa = 3.88AWVKK86 pKa = 8.55PVKK89 pKa = 9.53ITGEE93 pKa = 4.11VSVMSHH99 pKa = 5.65SGIYY103 pKa = 10.21DD104 pKa = 3.6GLTITPTHH112 pKa = 6.89IYY114 pKa = 10.58FSTKK118 pKa = 9.97YY119 pKa = 8.68LTAGSCEE126 pKa = 3.76VSYY129 pKa = 10.5EE130 pKa = 3.94YY131 pKa = 10.23STGKK135 pKa = 9.88SFHH138 pKa = 6.39VVGVHH143 pKa = 5.58TNAKK147 pKa = 9.52NSLYY151 pKa = 10.73VDD153 pKa = 4.01GVLVAEE159 pKa = 4.55TDD161 pKa = 3.59LTDD164 pKa = 3.45EE165 pKa = 4.24QQIDD169 pKa = 3.96SYY171 pKa = 12.04SFLTSDD177 pKa = 5.14LIAGQGSGTIALDD190 pKa = 3.54APAVYY195 pKa = 9.95TDD197 pKa = 3.73ALGAEE202 pKa = 4.95AISRR206 pKa = 11.84HH207 pKa = 5.2YY208 pKa = 10.94FNGVDD213 pKa = 3.35VDD215 pKa = 3.81TSEE218 pKa = 5.7AIAGFNDD225 pKa = 4.48AIFWNFTDD233 pKa = 3.48EE234 pKa = 4.21TRR236 pKa = 11.84NIAIKK241 pKa = 10.36KK242 pKa = 8.77VWASDD247 pKa = 4.22DD248 pKa = 3.7DD249 pKa = 3.81WSDD252 pKa = 3.52AVLTDD257 pKa = 3.2VAVIDD262 pKa = 4.07GTIVPSYY269 pKa = 10.41SQTEE273 pKa = 4.39TEE275 pKa = 4.26VVEE278 pKa = 5.07DD279 pKa = 4.01GLIVPVYY286 pKa = 10.23EE287 pKa = 4.19NTSLPGVWLASLSFTSIPEE306 pKa = 3.85EE307 pKa = 4.27TITDD311 pKa = 3.48ARR313 pKa = 11.84INWVGEE319 pKa = 3.63GSYY322 pKa = 10.12TIEE325 pKa = 4.06YY326 pKa = 10.3SVDD329 pKa = 3.25GGNIWLEE336 pKa = 4.16PSDD339 pKa = 4.15SGATVLDD346 pKa = 3.83NEE348 pKa = 4.55SVIDD352 pKa = 3.77IRR354 pKa = 11.84VTFDD358 pKa = 2.89GGIVDD363 pKa = 4.97DD364 pKa = 4.7EE365 pKa = 5.2SFVSSIEE372 pKa = 3.99VVGYY376 pKa = 9.97LDD378 pKa = 3.0KK379 pKa = 11.11YY380 pKa = 10.03YY381 pKa = 10.6RR382 pKa = 11.84GSRR385 pKa = 11.84ADD387 pKa = 3.39RR388 pKa = 11.84TAQMSGTGVVSSSYY402 pKa = 10.69FEE404 pKa = 4.97PIEE407 pKa = 4.0YY408 pKa = 10.74ADD410 pKa = 3.7VNGLRR415 pKa = 11.84IITGTVNVSLDD426 pKa = 3.37DD427 pKa = 4.06SYY429 pKa = 11.88EE430 pKa = 4.32GEE432 pKa = 4.43QEE434 pKa = 4.84AGDD437 pKa = 4.2LNVNGIDD444 pKa = 3.4MWIKK448 pKa = 8.59PTVGNILTATGISISRR464 pKa = 11.84VGNTIVFSGFSSFTVNGNSVTSGDD488 pKa = 3.71TVFDD492 pKa = 3.58SDD494 pKa = 3.26SWYY497 pKa = 10.53HH498 pKa = 5.08IAGVFSTPGNYY509 pKa = 10.43AMSIASAGTIISQLSVIYY527 pKa = 10.48GSLSLLGLQQIYY539 pKa = 10.17AAYY542 pKa = 10.17LGLPGLIVNDD552 pKa = 3.96TSTVDD557 pKa = 3.04IGEE560 pKa = 4.53GSPATNLYY568 pKa = 9.27AHH570 pKa = 6.59VWGITPAGG578 pKa = 3.48
MM1 pKa = 7.76SYY3 pKa = 10.53QLQVLTDD10 pKa = 3.56APFSYY15 pKa = 9.79WKK17 pKa = 10.51LDD19 pKa = 3.37EE20 pKa = 4.85TGPAFPDD27 pKa = 3.27SAGSMRR33 pKa = 11.84TADD36 pKa = 3.72LVGTITRR43 pKa = 11.84HH44 pKa = 5.3PALVTGSGNALVLDD58 pKa = 4.24NTNHH62 pKa = 7.1LDD64 pKa = 3.3MDD66 pKa = 4.3DD67 pKa = 3.64PVFNKK72 pKa = 10.34GYY74 pKa = 8.92EE75 pKa = 3.89LRR77 pKa = 11.84QFSLEE82 pKa = 3.88AWVKK86 pKa = 8.55PVKK89 pKa = 9.53ITGEE93 pKa = 4.11VSVMSHH99 pKa = 5.65SGIYY103 pKa = 10.21DD104 pKa = 3.6GLTITPTHH112 pKa = 6.89IYY114 pKa = 10.58FSTKK118 pKa = 9.97YY119 pKa = 8.68LTAGSCEE126 pKa = 3.76VSYY129 pKa = 10.5EE130 pKa = 3.94YY131 pKa = 10.23STGKK135 pKa = 9.88SFHH138 pKa = 6.39VVGVHH143 pKa = 5.58TNAKK147 pKa = 9.52NSLYY151 pKa = 10.73VDD153 pKa = 4.01GVLVAEE159 pKa = 4.55TDD161 pKa = 3.59LTDD164 pKa = 3.45EE165 pKa = 4.24QQIDD169 pKa = 3.96SYY171 pKa = 12.04SFLTSDD177 pKa = 5.14LIAGQGSGTIALDD190 pKa = 3.54APAVYY195 pKa = 9.95TDD197 pKa = 3.73ALGAEE202 pKa = 4.95AISRR206 pKa = 11.84HH207 pKa = 5.2YY208 pKa = 10.94FNGVDD213 pKa = 3.35VDD215 pKa = 3.81TSEE218 pKa = 5.7AIAGFNDD225 pKa = 4.48AIFWNFTDD233 pKa = 3.48EE234 pKa = 4.21TRR236 pKa = 11.84NIAIKK241 pKa = 10.36KK242 pKa = 8.77VWASDD247 pKa = 4.22DD248 pKa = 3.7DD249 pKa = 3.81WSDD252 pKa = 3.52AVLTDD257 pKa = 3.2VAVIDD262 pKa = 4.07GTIVPSYY269 pKa = 10.41SQTEE273 pKa = 4.39TEE275 pKa = 4.26VVEE278 pKa = 5.07DD279 pKa = 4.01GLIVPVYY286 pKa = 10.23EE287 pKa = 4.19NTSLPGVWLASLSFTSIPEE306 pKa = 3.85EE307 pKa = 4.27TITDD311 pKa = 3.48ARR313 pKa = 11.84INWVGEE319 pKa = 3.63GSYY322 pKa = 10.12TIEE325 pKa = 4.06YY326 pKa = 10.3SVDD329 pKa = 3.25GGNIWLEE336 pKa = 4.16PSDD339 pKa = 4.15SGATVLDD346 pKa = 3.83NEE348 pKa = 4.55SVIDD352 pKa = 3.77IRR354 pKa = 11.84VTFDD358 pKa = 2.89GGIVDD363 pKa = 4.97DD364 pKa = 4.7EE365 pKa = 5.2SFVSSIEE372 pKa = 3.99VVGYY376 pKa = 9.97LDD378 pKa = 3.0KK379 pKa = 11.11YY380 pKa = 10.03YY381 pKa = 10.6RR382 pKa = 11.84GSRR385 pKa = 11.84ADD387 pKa = 3.39RR388 pKa = 11.84TAQMSGTGVVSSSYY402 pKa = 10.69FEE404 pKa = 4.97PIEE407 pKa = 4.0YY408 pKa = 10.74ADD410 pKa = 3.7VNGLRR415 pKa = 11.84IITGTVNVSLDD426 pKa = 3.37DD427 pKa = 4.06SYY429 pKa = 11.88EE430 pKa = 4.32GEE432 pKa = 4.43QEE434 pKa = 4.84AGDD437 pKa = 4.2LNVNGIDD444 pKa = 3.4MWIKK448 pKa = 8.59PTVGNILTATGISISRR464 pKa = 11.84VGNTIVFSGFSSFTVNGNSVTSGDD488 pKa = 3.71TVFDD492 pKa = 3.58SDD494 pKa = 3.26SWYY497 pKa = 10.53HH498 pKa = 5.08IAGVFSTPGNYY509 pKa = 10.43AMSIASAGTIISQLSVIYY527 pKa = 10.48GSLSLLGLQQIYY539 pKa = 10.17AAYY542 pKa = 10.17LGLPGLIVNDD552 pKa = 3.96TSTVDD557 pKa = 3.04IGEE560 pKa = 4.53GSPATNLYY568 pKa = 9.27AHH570 pKa = 6.59VWGITPAGG578 pKa = 3.48
Molecular weight: 61.96 kDa
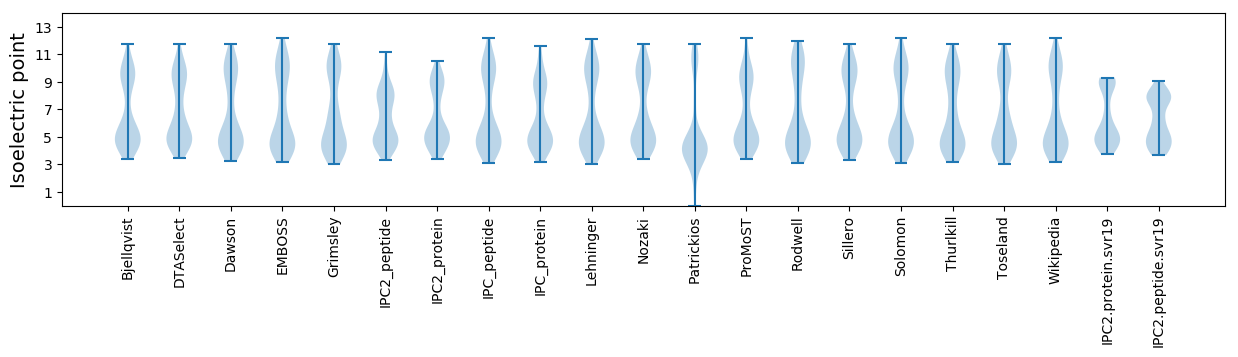
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0RT10|A0A0A0RT10_9CAUD Uncharacterized protein OS=Streptomyces phage Jay2Jay OX=1556290 GN=51 PE=4 SV=1
MM1 pKa = 7.15KK2 pKa = 9.78TDD4 pKa = 3.35EE5 pKa = 4.86DD6 pKa = 3.93RR7 pKa = 11.84CIEE10 pKa = 4.13YY11 pKa = 10.35GVPYY15 pKa = 9.83DD16 pKa = 3.79PSITRR21 pKa = 11.84PVVYY25 pKa = 10.03AQSNWTCHH33 pKa = 5.64LCGKK37 pKa = 9.81RR38 pKa = 11.84VSKK41 pKa = 10.3RR42 pKa = 11.84LKK44 pKa = 9.48YY45 pKa = 8.65PHH47 pKa = 7.26PKK49 pKa = 9.85SASLDD54 pKa = 3.89HH55 pKa = 6.64IVPLSWRR62 pKa = 11.84KK63 pKa = 9.93DD64 pKa = 3.68SPGHH68 pKa = 4.45VWGNVALAHH77 pKa = 6.12LRR79 pKa = 11.84CNQSKK84 pKa = 9.12GARR87 pKa = 11.84FAGSTRR93 pKa = 11.84PAPRR97 pKa = 11.84RR98 pKa = 11.84PGFINNVWKK107 pKa = 10.6LRR109 pKa = 11.84IALFGFTGAMFYY121 pKa = 10.71FGATPLALTIAVALCILSVIKK142 pKa = 10.51VKK144 pKa = 10.53KK145 pKa = 8.25SRR147 pKa = 11.84RR148 pKa = 11.84RR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84RR152 pKa = 11.84AWWKK156 pKa = 10.21LL157 pKa = 3.23
MM1 pKa = 7.15KK2 pKa = 9.78TDD4 pKa = 3.35EE5 pKa = 4.86DD6 pKa = 3.93RR7 pKa = 11.84CIEE10 pKa = 4.13YY11 pKa = 10.35GVPYY15 pKa = 9.83DD16 pKa = 3.79PSITRR21 pKa = 11.84PVVYY25 pKa = 10.03AQSNWTCHH33 pKa = 5.64LCGKK37 pKa = 9.81RR38 pKa = 11.84VSKK41 pKa = 10.3RR42 pKa = 11.84LKK44 pKa = 9.48YY45 pKa = 8.65PHH47 pKa = 7.26PKK49 pKa = 9.85SASLDD54 pKa = 3.89HH55 pKa = 6.64IVPLSWRR62 pKa = 11.84KK63 pKa = 9.93DD64 pKa = 3.68SPGHH68 pKa = 4.45VWGNVALAHH77 pKa = 6.12LRR79 pKa = 11.84CNQSKK84 pKa = 9.12GARR87 pKa = 11.84FAGSTRR93 pKa = 11.84PAPRR97 pKa = 11.84RR98 pKa = 11.84PGFINNVWKK107 pKa = 10.6LRR109 pKa = 11.84IALFGFTGAMFYY121 pKa = 10.71FGATPLALTIAVALCILSVIKK142 pKa = 10.51VKK144 pKa = 10.53KK145 pKa = 8.25SRR147 pKa = 11.84RR148 pKa = 11.84RR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84RR152 pKa = 11.84AWWKK156 pKa = 10.21LL157 pKa = 3.23
Molecular weight: 17.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
34582 |
23 |
2103 |
165.5 |
18.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.444 ± 0.379 | 0.992 ± 0.11 |
6.48 ± 0.179 | 6.833 ± 0.299 |
3.831 ± 0.12 | 7.504 ± 0.177 |
1.877 ± 0.128 | 5.384 ± 0.149 |
6.252 ± 0.278 | 7.226 ± 0.198 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.903 ± 0.118 | 4.572 ± 0.15 |
3.803 ± 0.177 | 3.279 ± 0.186 |
5.584 ± 0.182 | 6.13 ± 0.248 |
6.171 ± 0.314 | 7.145 ± 0.209 |
1.885 ± 0.094 | 3.704 ± 0.182 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
