
Beihai shrimp virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
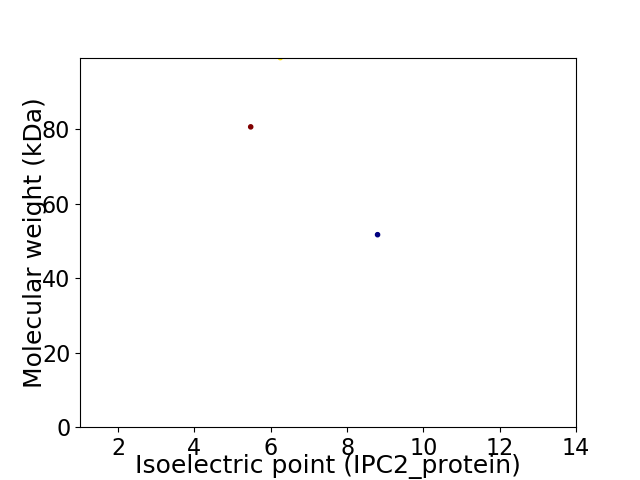
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
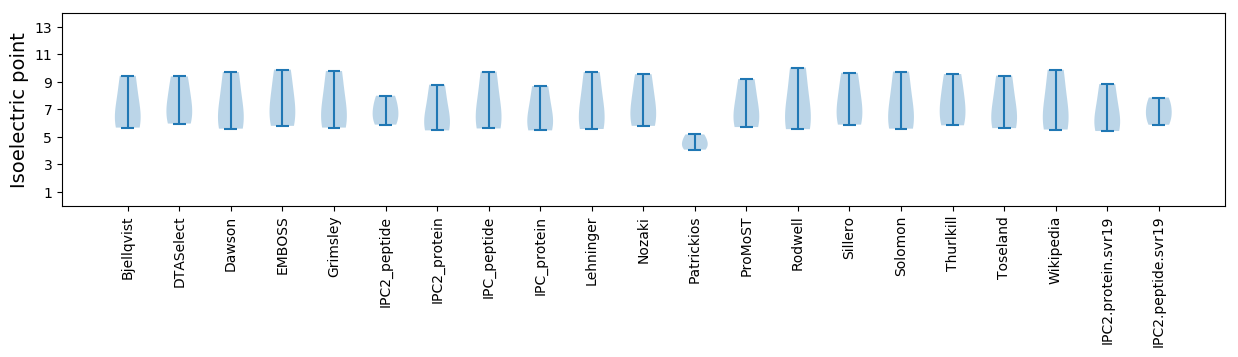
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE77|A0A1L3KE77_9VIRU Uncharacterized protein OS=Beihai shrimp virus 5 OX=1922671 PE=4 SV=1
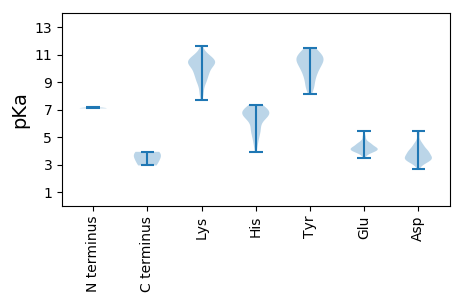
MM1 pKa = 7.1GRR3 pKa = 11.84EE4 pKa = 3.99LEE6 pKa = 4.26TRR8 pKa = 11.84SDD10 pKa = 3.87GSGTTNGPNKK20 pKa = 9.56FRR22 pKa = 11.84ACLDD26 pKa = 3.47TTHH29 pKa = 6.39QGSVFSTFGNSKK41 pKa = 10.1IGVSNLGHH49 pKa = 6.31AVSPAKK55 pKa = 10.4EE56 pKa = 3.87EE57 pKa = 4.24SGVLDD62 pKa = 3.29SRR64 pKa = 11.84GSGNVDD70 pKa = 2.69PWSATQRR77 pKa = 11.84TTGSICVFDD86 pKa = 4.36GNYY89 pKa = 10.15GGCEE93 pKa = 3.7TSTSGTYY100 pKa = 9.68RR101 pKa = 11.84EE102 pKa = 4.1QQRR105 pKa = 11.84IVYY108 pKa = 9.58EE109 pKa = 3.95SGTIGKK115 pKa = 9.71RR116 pKa = 11.84KK117 pKa = 9.1IEE119 pKa = 3.93RR120 pKa = 11.84FEE122 pKa = 4.04SCEE125 pKa = 3.75RR126 pKa = 11.84RR127 pKa = 11.84AGTSQDD133 pKa = 3.61NEE135 pKa = 4.34LKK137 pKa = 10.5NSEE140 pKa = 4.48NVGEE144 pKa = 4.4TCDD147 pKa = 3.49GRR149 pKa = 11.84QIPTRR154 pKa = 11.84WFGKK158 pKa = 10.11SEE160 pKa = 4.11TDD162 pKa = 3.2EE163 pKa = 4.4PTSSGQIGNASSCEE177 pKa = 3.89EE178 pKa = 3.95SSEE181 pKa = 4.49SFGTRR186 pKa = 11.84TTEE189 pKa = 3.93RR190 pKa = 11.84RR191 pKa = 11.84NDD193 pKa = 3.32GCQSTGCASIAQEE206 pKa = 4.0TTTQQEE212 pKa = 4.52TEE214 pKa = 4.3FVSDD218 pKa = 3.94CSSEE222 pKa = 3.91RR223 pKa = 11.84RR224 pKa = 11.84SSIGSPCGFAPAGDD238 pKa = 4.15GEE240 pKa = 5.1PIFARR245 pKa = 11.84RR246 pKa = 11.84DD247 pKa = 3.43QPTMGRR253 pKa = 11.84FAWLHH258 pKa = 4.94EE259 pKa = 4.16QAVSAGCEE267 pKa = 3.99PIADD271 pKa = 4.21KK272 pKa = 10.4STSSAFEE279 pKa = 3.83WPPYY283 pKa = 10.56GGDD286 pKa = 3.41SEE288 pKa = 4.54YY289 pKa = 11.16QSLCAHH295 pKa = 6.88AKK297 pKa = 9.85LRR299 pKa = 11.84ADD301 pKa = 3.09AHH303 pKa = 6.6SNARR307 pKa = 11.84TPTKK311 pKa = 10.44LEE313 pKa = 3.65RR314 pKa = 11.84SAAIAATYY322 pKa = 9.27RR323 pKa = 11.84ANSKK327 pKa = 9.68FSKK330 pKa = 9.86WSLPVDD336 pKa = 4.58FLSRR340 pKa = 11.84AHH342 pKa = 6.59LVRR345 pKa = 11.84VIRR348 pKa = 11.84ALDD351 pKa = 3.73FTSSPGLPICKK362 pKa = 9.58YY363 pKa = 10.47SPTISDD369 pKa = 4.35FLRR372 pKa = 11.84VDD374 pKa = 3.88PVTGLPDD381 pKa = 3.25RR382 pKa = 11.84EE383 pKa = 4.08RR384 pKa = 11.84EE385 pKa = 3.85DD386 pKa = 3.87RR387 pKa = 11.84LIEE390 pKa = 4.08EE391 pKa = 4.54VYY393 pKa = 10.3RR394 pKa = 11.84WLDD397 pKa = 3.35EE398 pKa = 4.71GDD400 pKa = 2.96IQYY403 pKa = 10.68YY404 pKa = 9.23IRR406 pKa = 11.84LFIKK410 pKa = 10.42YY411 pKa = 8.61EE412 pKa = 3.7PHH414 pKa = 6.57KK415 pKa = 10.45LSKK418 pKa = 10.46ISAKK422 pKa = 9.97RR423 pKa = 11.84FRR425 pKa = 11.84LIWSIPLVHH434 pKa = 6.71QIVDD438 pKa = 3.55HH439 pKa = 6.74LLFDD443 pKa = 3.99AQNDD447 pKa = 4.06RR448 pKa = 11.84EE449 pKa = 4.4VQTSSLSEE457 pKa = 4.0FKK459 pKa = 10.86GGSSFAKK466 pKa = 10.21GGCLDD471 pKa = 4.19YY472 pKa = 10.94FSHH475 pKa = 6.26TPGWHH480 pKa = 6.06LSADD484 pKa = 3.24KK485 pKa = 10.96SLWDD489 pKa = 3.54FTVPAWACEE498 pKa = 3.75DD499 pKa = 3.72DD500 pKa = 4.45LEE502 pKa = 4.49FRR504 pKa = 11.84SMCCLDD510 pKa = 3.35SALNPEE516 pKa = 4.61KK517 pKa = 10.62FKK519 pKa = 10.61IWLEE523 pKa = 4.06CAQRR527 pKa = 11.84RR528 pKa = 11.84YY529 pKa = 11.04NEE531 pKa = 4.42MFNLDD536 pKa = 3.84YY537 pKa = 10.85DD538 pKa = 4.52VKK540 pKa = 11.04SKK542 pKa = 9.05PTLIQLSDD550 pKa = 3.24GRR552 pKa = 11.84RR553 pKa = 11.84FLQLTKK559 pKa = 11.04GLMKK563 pKa = 10.25SGSVITLSANSHH575 pKa = 4.91MQSFFHH581 pKa = 6.9RR582 pKa = 11.84IAWSRR587 pKa = 11.84VSDD590 pKa = 4.01VPAPLIWSMGDD601 pKa = 3.27DD602 pKa = 3.24TTLRR606 pKa = 11.84VSRR609 pKa = 11.84DD610 pKa = 3.55FPTEE614 pKa = 3.81LYY616 pKa = 10.7LRR618 pKa = 11.84EE619 pKa = 4.0TSEE622 pKa = 3.97LGAIVKK628 pKa = 8.46FANLTVNEE636 pKa = 4.2YY637 pKa = 9.98EE638 pKa = 4.13FAGMIIKK645 pKa = 10.15QPMCLIPSKK654 pKa = 11.13NDD656 pKa = 2.82THH658 pKa = 6.74YY659 pKa = 11.36YY660 pKa = 10.28KK661 pKa = 11.11LLFEE665 pKa = 4.32TDD667 pKa = 3.08EE668 pKa = 4.61RR669 pKa = 11.84LLEE672 pKa = 4.14MLHH675 pKa = 6.8SYY677 pKa = 10.31QFLYY681 pKa = 10.88AADD684 pKa = 4.02NRR686 pKa = 11.84LKK688 pKa = 9.97TIQYY692 pKa = 9.89VLSLLPGGSEE702 pKa = 4.11RR703 pKa = 11.84IEE705 pKa = 3.9TPMYY709 pKa = 9.91LNQWLTEE716 pKa = 4.32PGSSS720 pKa = 3.57
MM1 pKa = 7.1GRR3 pKa = 11.84EE4 pKa = 3.99LEE6 pKa = 4.26TRR8 pKa = 11.84SDD10 pKa = 3.87GSGTTNGPNKK20 pKa = 9.56FRR22 pKa = 11.84ACLDD26 pKa = 3.47TTHH29 pKa = 6.39QGSVFSTFGNSKK41 pKa = 10.1IGVSNLGHH49 pKa = 6.31AVSPAKK55 pKa = 10.4EE56 pKa = 3.87EE57 pKa = 4.24SGVLDD62 pKa = 3.29SRR64 pKa = 11.84GSGNVDD70 pKa = 2.69PWSATQRR77 pKa = 11.84TTGSICVFDD86 pKa = 4.36GNYY89 pKa = 10.15GGCEE93 pKa = 3.7TSTSGTYY100 pKa = 9.68RR101 pKa = 11.84EE102 pKa = 4.1QQRR105 pKa = 11.84IVYY108 pKa = 9.58EE109 pKa = 3.95SGTIGKK115 pKa = 9.71RR116 pKa = 11.84KK117 pKa = 9.1IEE119 pKa = 3.93RR120 pKa = 11.84FEE122 pKa = 4.04SCEE125 pKa = 3.75RR126 pKa = 11.84RR127 pKa = 11.84AGTSQDD133 pKa = 3.61NEE135 pKa = 4.34LKK137 pKa = 10.5NSEE140 pKa = 4.48NVGEE144 pKa = 4.4TCDD147 pKa = 3.49GRR149 pKa = 11.84QIPTRR154 pKa = 11.84WFGKK158 pKa = 10.11SEE160 pKa = 4.11TDD162 pKa = 3.2EE163 pKa = 4.4PTSSGQIGNASSCEE177 pKa = 3.89EE178 pKa = 3.95SSEE181 pKa = 4.49SFGTRR186 pKa = 11.84TTEE189 pKa = 3.93RR190 pKa = 11.84RR191 pKa = 11.84NDD193 pKa = 3.32GCQSTGCASIAQEE206 pKa = 4.0TTTQQEE212 pKa = 4.52TEE214 pKa = 4.3FVSDD218 pKa = 3.94CSSEE222 pKa = 3.91RR223 pKa = 11.84RR224 pKa = 11.84SSIGSPCGFAPAGDD238 pKa = 4.15GEE240 pKa = 5.1PIFARR245 pKa = 11.84RR246 pKa = 11.84DD247 pKa = 3.43QPTMGRR253 pKa = 11.84FAWLHH258 pKa = 4.94EE259 pKa = 4.16QAVSAGCEE267 pKa = 3.99PIADD271 pKa = 4.21KK272 pKa = 10.4STSSAFEE279 pKa = 3.83WPPYY283 pKa = 10.56GGDD286 pKa = 3.41SEE288 pKa = 4.54YY289 pKa = 11.16QSLCAHH295 pKa = 6.88AKK297 pKa = 9.85LRR299 pKa = 11.84ADD301 pKa = 3.09AHH303 pKa = 6.6SNARR307 pKa = 11.84TPTKK311 pKa = 10.44LEE313 pKa = 3.65RR314 pKa = 11.84SAAIAATYY322 pKa = 9.27RR323 pKa = 11.84ANSKK327 pKa = 9.68FSKK330 pKa = 9.86WSLPVDD336 pKa = 4.58FLSRR340 pKa = 11.84AHH342 pKa = 6.59LVRR345 pKa = 11.84VIRR348 pKa = 11.84ALDD351 pKa = 3.73FTSSPGLPICKK362 pKa = 9.58YY363 pKa = 10.47SPTISDD369 pKa = 4.35FLRR372 pKa = 11.84VDD374 pKa = 3.88PVTGLPDD381 pKa = 3.25RR382 pKa = 11.84EE383 pKa = 4.08RR384 pKa = 11.84EE385 pKa = 3.85DD386 pKa = 3.87RR387 pKa = 11.84LIEE390 pKa = 4.08EE391 pKa = 4.54VYY393 pKa = 10.3RR394 pKa = 11.84WLDD397 pKa = 3.35EE398 pKa = 4.71GDD400 pKa = 2.96IQYY403 pKa = 10.68YY404 pKa = 9.23IRR406 pKa = 11.84LFIKK410 pKa = 10.42YY411 pKa = 8.61EE412 pKa = 3.7PHH414 pKa = 6.57KK415 pKa = 10.45LSKK418 pKa = 10.46ISAKK422 pKa = 9.97RR423 pKa = 11.84FRR425 pKa = 11.84LIWSIPLVHH434 pKa = 6.71QIVDD438 pKa = 3.55HH439 pKa = 6.74LLFDD443 pKa = 3.99AQNDD447 pKa = 4.06RR448 pKa = 11.84EE449 pKa = 4.4VQTSSLSEE457 pKa = 4.0FKK459 pKa = 10.86GGSSFAKK466 pKa = 10.21GGCLDD471 pKa = 4.19YY472 pKa = 10.94FSHH475 pKa = 6.26TPGWHH480 pKa = 6.06LSADD484 pKa = 3.24KK485 pKa = 10.96SLWDD489 pKa = 3.54FTVPAWACEE498 pKa = 3.75DD499 pKa = 3.72DD500 pKa = 4.45LEE502 pKa = 4.49FRR504 pKa = 11.84SMCCLDD510 pKa = 3.35SALNPEE516 pKa = 4.61KK517 pKa = 10.62FKK519 pKa = 10.61IWLEE523 pKa = 4.06CAQRR527 pKa = 11.84RR528 pKa = 11.84YY529 pKa = 11.04NEE531 pKa = 4.42MFNLDD536 pKa = 3.84YY537 pKa = 10.85DD538 pKa = 4.52VKK540 pKa = 11.04SKK542 pKa = 9.05PTLIQLSDD550 pKa = 3.24GRR552 pKa = 11.84RR553 pKa = 11.84FLQLTKK559 pKa = 11.04GLMKK563 pKa = 10.25SGSVITLSANSHH575 pKa = 4.91MQSFFHH581 pKa = 6.9RR582 pKa = 11.84IAWSRR587 pKa = 11.84VSDD590 pKa = 4.01VPAPLIWSMGDD601 pKa = 3.27DD602 pKa = 3.24TTLRR606 pKa = 11.84VSRR609 pKa = 11.84DD610 pKa = 3.55FPTEE614 pKa = 3.81LYY616 pKa = 10.7LRR618 pKa = 11.84EE619 pKa = 4.0TSEE622 pKa = 3.97LGAIVKK628 pKa = 8.46FANLTVNEE636 pKa = 4.2YY637 pKa = 9.98EE638 pKa = 4.13FAGMIIKK645 pKa = 10.15QPMCLIPSKK654 pKa = 11.13NDD656 pKa = 2.82THH658 pKa = 6.74YY659 pKa = 11.36YY660 pKa = 10.28KK661 pKa = 11.11LLFEE665 pKa = 4.32TDD667 pKa = 3.08EE668 pKa = 4.61RR669 pKa = 11.84LLEE672 pKa = 4.14MLHH675 pKa = 6.8SYY677 pKa = 10.31QFLYY681 pKa = 10.88AADD684 pKa = 4.02NRR686 pKa = 11.84LKK688 pKa = 9.97TIQYY692 pKa = 9.89VLSLLPGGSEE702 pKa = 4.11RR703 pKa = 11.84IEE705 pKa = 3.9TPMYY709 pKa = 9.91LNQWLTEE716 pKa = 4.32PGSSS720 pKa = 3.57
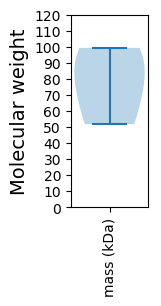
Molecular weight: 80.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE78|A0A1L3KE78_9VIRU Uncharacterized protein OS=Beihai shrimp virus 5 OX=1922671 PE=4 SV=1
MM1 pKa = 7.15QPNGTVTVSTKK12 pKa = 9.03NKK14 pKa = 8.9KK15 pKa = 9.17RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84QKK20 pKa = 10.51RR21 pKa = 11.84KK22 pKa = 8.6NKK24 pKa = 8.84TVIQVVQKK32 pKa = 9.83PKK34 pKa = 8.29RR35 pKa = 11.84QRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84GLALNQAPVAAGPVTRR56 pKa = 11.84TTFTGPLITSGMDD69 pKa = 3.26YY70 pKa = 11.13LDD72 pKa = 5.18ADD74 pKa = 5.44LIPAKK79 pKa = 10.51VDD81 pKa = 3.44DD82 pKa = 4.27GTLIVDD88 pKa = 3.84KK89 pKa = 10.7LLNPSQLPGTHH100 pKa = 6.75FSAIAGGFDD109 pKa = 3.17RR110 pKa = 11.84YY111 pKa = 10.61QFTSLSVQVISRR123 pKa = 11.84VPTTVGGGYY132 pKa = 9.38IAALSSDD139 pKa = 3.39VVEE142 pKa = 4.23EE143 pKa = 4.28FEE145 pKa = 5.03NGVSLKK151 pKa = 9.4QHH153 pKa = 4.43TMAMEE158 pKa = 4.98GSKK161 pKa = 10.46GAPWWTSVSVTMGKK175 pKa = 7.73QTGEE179 pKa = 3.58WYY181 pKa = 10.85YY182 pKa = 11.5NSPQSNEE189 pKa = 3.8RR190 pKa = 11.84FKK192 pKa = 9.01TTQARR197 pKa = 11.84FLMVVDD203 pKa = 5.32GPPTNLSMNSSVQVTLLLKK222 pKa = 9.99WKK224 pKa = 10.42AKK226 pKa = 9.15FASPTGMAPRR236 pKa = 11.84TVQKK240 pKa = 8.91FTIPRR245 pKa = 11.84GTVFTSPNPTHH256 pKa = 7.27PYY258 pKa = 8.16YY259 pKa = 10.86GQYY262 pKa = 10.76LYY264 pKa = 11.37VPGTLSEE271 pKa = 4.08FWKK274 pKa = 10.56ASKK277 pKa = 10.39FSYY280 pKa = 10.57VYY282 pKa = 9.99IVVPGAEE289 pKa = 4.5YY290 pKa = 10.76KK291 pKa = 9.1RR292 pKa = 11.84TPPAEE297 pKa = 4.17SANVFAIQVGKK308 pKa = 8.98TSEE311 pKa = 4.25GSWTIGLWEE320 pKa = 4.56TIEE323 pKa = 4.19SCEE326 pKa = 4.43KK327 pKa = 10.7VDD329 pKa = 4.83FNAPGGRR336 pKa = 11.84SGILCNGDD344 pKa = 2.66WTLDD348 pKa = 3.32RR349 pKa = 11.84DD350 pKa = 3.56VDD352 pKa = 4.3FRR354 pKa = 11.84PLCPVTPKK362 pKa = 10.29KK363 pKa = 10.27RR364 pKa = 11.84SYY366 pKa = 11.46LNLSVSDD373 pKa = 4.39DD374 pKa = 3.93FTHH377 pKa = 7.01KK378 pKa = 10.57AARR381 pKa = 11.84NLINTEE387 pKa = 3.9RR388 pKa = 11.84EE389 pKa = 4.41SIIHH393 pKa = 7.0ADD395 pKa = 3.65AIGNRR400 pKa = 11.84SNKK403 pKa = 9.48LLQASTEE410 pKa = 4.0LLQNQMKK417 pKa = 7.97MTEE420 pKa = 4.23KK421 pKa = 10.38LSSLMEE427 pKa = 4.43SLTLTIPIIGTTLRR441 pKa = 11.84TMNVEE446 pKa = 4.19VPIPPASSSCDD457 pKa = 3.11DD458 pKa = 3.47STFSVVAEE466 pKa = 4.06EE467 pKa = 4.62TEE469 pKa = 3.92
MM1 pKa = 7.15QPNGTVTVSTKK12 pKa = 9.03NKK14 pKa = 8.9KK15 pKa = 9.17RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84QKK20 pKa = 10.51RR21 pKa = 11.84KK22 pKa = 8.6NKK24 pKa = 8.84TVIQVVQKK32 pKa = 9.83PKK34 pKa = 8.29RR35 pKa = 11.84QRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84GLALNQAPVAAGPVTRR56 pKa = 11.84TTFTGPLITSGMDD69 pKa = 3.26YY70 pKa = 11.13LDD72 pKa = 5.18ADD74 pKa = 5.44LIPAKK79 pKa = 10.51VDD81 pKa = 3.44DD82 pKa = 4.27GTLIVDD88 pKa = 3.84KK89 pKa = 10.7LLNPSQLPGTHH100 pKa = 6.75FSAIAGGFDD109 pKa = 3.17RR110 pKa = 11.84YY111 pKa = 10.61QFTSLSVQVISRR123 pKa = 11.84VPTTVGGGYY132 pKa = 9.38IAALSSDD139 pKa = 3.39VVEE142 pKa = 4.23EE143 pKa = 4.28FEE145 pKa = 5.03NGVSLKK151 pKa = 9.4QHH153 pKa = 4.43TMAMEE158 pKa = 4.98GSKK161 pKa = 10.46GAPWWTSVSVTMGKK175 pKa = 7.73QTGEE179 pKa = 3.58WYY181 pKa = 10.85YY182 pKa = 11.5NSPQSNEE189 pKa = 3.8RR190 pKa = 11.84FKK192 pKa = 9.01TTQARR197 pKa = 11.84FLMVVDD203 pKa = 5.32GPPTNLSMNSSVQVTLLLKK222 pKa = 9.99WKK224 pKa = 10.42AKK226 pKa = 9.15FASPTGMAPRR236 pKa = 11.84TVQKK240 pKa = 8.91FTIPRR245 pKa = 11.84GTVFTSPNPTHH256 pKa = 7.27PYY258 pKa = 8.16YY259 pKa = 10.86GQYY262 pKa = 10.76LYY264 pKa = 11.37VPGTLSEE271 pKa = 4.08FWKK274 pKa = 10.56ASKK277 pKa = 10.39FSYY280 pKa = 10.57VYY282 pKa = 9.99IVVPGAEE289 pKa = 4.5YY290 pKa = 10.76KK291 pKa = 9.1RR292 pKa = 11.84TPPAEE297 pKa = 4.17SANVFAIQVGKK308 pKa = 8.98TSEE311 pKa = 4.25GSWTIGLWEE320 pKa = 4.56TIEE323 pKa = 4.19SCEE326 pKa = 4.43KK327 pKa = 10.7VDD329 pKa = 4.83FNAPGGRR336 pKa = 11.84SGILCNGDD344 pKa = 2.66WTLDD348 pKa = 3.32RR349 pKa = 11.84DD350 pKa = 3.56VDD352 pKa = 4.3FRR354 pKa = 11.84PLCPVTPKK362 pKa = 10.29KK363 pKa = 10.27RR364 pKa = 11.84SYY366 pKa = 11.46LNLSVSDD373 pKa = 4.39DD374 pKa = 3.93FTHH377 pKa = 7.01KK378 pKa = 10.57AARR381 pKa = 11.84NLINTEE387 pKa = 3.9RR388 pKa = 11.84EE389 pKa = 4.41SIIHH393 pKa = 7.0ADD395 pKa = 3.65AIGNRR400 pKa = 11.84SNKK403 pKa = 9.48LLQASTEE410 pKa = 4.0LLQNQMKK417 pKa = 7.97MTEE420 pKa = 4.23KK421 pKa = 10.38LSSLMEE427 pKa = 4.43SLTLTIPIIGTTLRR441 pKa = 11.84TMNVEE446 pKa = 4.19VPIPPASSSCDD457 pKa = 3.11DD458 pKa = 3.47STFSVVAEE466 pKa = 4.06EE467 pKa = 4.62TEE469 pKa = 3.92
Molecular weight: 51.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2066 |
469 |
877 |
688.7 |
77.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.002 ± 0.098 | 1.597 ± 0.408 |
5.131 ± 0.379 | 6.873 ± 0.742 |
4.211 ± 0.23 | 6.05 ± 0.685 |
2.033 ± 0.351 | 5.227 ± 0.424 |
6.196 ± 0.817 | 8.132 ± 0.265 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.372 ± 0.338 | 3.872 ± 0.277 |
5.663 ± 0.44 | 4.55 ± 0.462 |
5.566 ± 0.56 | 8.422 ± 1.32 |
7.406 ± 0.824 | 6.583 ± 1.038 |
1.404 ± 0.301 | 2.711 ± 0.16 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
