
Gammapapillomavirus 24
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
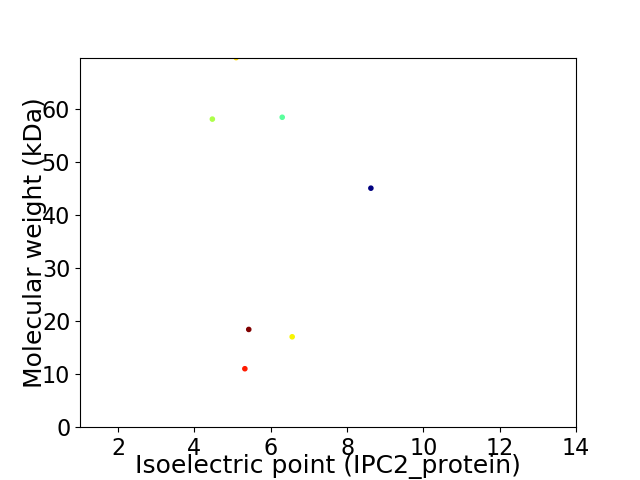
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2AM75|A0A2D2AM75_9PAPI Replication protein E1 OS=Gammapapillomavirus 24 OX=1961681 GN=E1 PE=3 SV=1
MM1 pKa = 7.62EE2 pKa = 5.41PRR4 pKa = 11.84AKK6 pKa = 9.9RR7 pKa = 11.84QKK9 pKa = 10.17RR10 pKa = 11.84DD11 pKa = 3.45SVEE14 pKa = 3.53NLYY17 pKa = 10.18RR18 pKa = 11.84QCKK21 pKa = 8.4LTGQCPDD28 pKa = 3.27DD29 pKa = 4.54VINKK33 pKa = 9.42VEE35 pKa = 4.15GNTLADD41 pKa = 3.99RR42 pKa = 11.84LLKK45 pKa = 10.5ILGSVIYY52 pKa = 10.6LGGLGIGTGRR62 pKa = 11.84GSGGSTGYY70 pKa = 10.46RR71 pKa = 11.84PIGATTPRR79 pKa = 11.84VTDD82 pKa = 3.47STPIRR87 pKa = 11.84PTVPVDD93 pKa = 3.3PLGPTDD99 pKa = 4.24VLPVDD104 pKa = 3.63PAGPSIVPLTEE115 pKa = 3.78SGIPEE120 pKa = 3.91DD121 pKa = 4.43TIIEE125 pKa = 4.33TSGTAVTVNPTEE137 pKa = 4.19PPVITNIDD145 pKa = 4.31PISDD149 pKa = 3.57VTGVEE154 pKa = 4.11GQPTIITGEE163 pKa = 4.16DD164 pKa = 3.31DD165 pKa = 3.69SVAVLDD171 pKa = 4.36VQPSTPAPKK180 pKa = 9.96RR181 pKa = 11.84VTIGVRR187 pKa = 11.84PRR189 pKa = 11.84TSSPSEE195 pKa = 3.62PTLYY199 pKa = 10.2PIPEE203 pKa = 4.14PAAPDD208 pKa = 3.34FTVFVDD214 pKa = 4.21AQAVGEE220 pKa = 4.53TIGGEE225 pKa = 4.15YY226 pKa = 9.98IPLEE230 pKa = 4.4EE231 pKa = 5.01FSQIQNFEE239 pKa = 3.9IEE241 pKa = 4.32EE242 pKa = 4.05QQTLSSTPEE251 pKa = 3.53QRR253 pKa = 11.84LNRR256 pKa = 11.84AFQRR260 pKa = 11.84ARR262 pKa = 11.84DD263 pKa = 3.75LYY265 pKa = 10.82NRR267 pKa = 11.84QITQIRR273 pKa = 11.84TRR275 pKa = 11.84NLDD278 pKa = 3.42FLGAASRR285 pKa = 11.84AVQFEE290 pKa = 4.21FEE292 pKa = 4.35NPAFVDD298 pKa = 4.1DD299 pKa = 3.74VTLQFEE305 pKa = 4.18QDD307 pKa = 3.6VRR309 pKa = 11.84DD310 pKa = 3.84VAAAPDD316 pKa = 3.96PDD318 pKa = 3.57FRR320 pKa = 11.84DD321 pKa = 3.0IVTLHH326 pKa = 6.93RR327 pKa = 11.84ARR329 pKa = 11.84YY330 pKa = 7.11STTDD334 pKa = 2.98TGRR337 pKa = 11.84VRR339 pKa = 11.84VSRR342 pKa = 11.84LGKK345 pKa = 9.98RR346 pKa = 11.84GTIVTRR352 pKa = 11.84SGAQIGEE359 pKa = 4.3NVHH362 pKa = 6.74FYY364 pKa = 11.29YY365 pKa = 10.63DD366 pKa = 3.37VSTIGSDD373 pKa = 3.03AADD376 pKa = 4.67AIEE379 pKa = 4.85LSTIGQHH386 pKa = 6.19SGDD389 pKa = 3.93SAIVDD394 pKa = 3.93ALAEE398 pKa = 4.21SSFINPIEE406 pKa = 4.54NPDD409 pKa = 3.5TTYY412 pKa = 11.17TEE414 pKa = 4.76ADD416 pKa = 3.53LLDD419 pKa = 3.98TQVEE423 pKa = 4.44DD424 pKa = 4.06FDD426 pKa = 4.71GLHH429 pKa = 6.6LVLTSSGRR437 pKa = 11.84RR438 pKa = 11.84GDD440 pKa = 3.77VYY442 pKa = 10.94SIPTLPPGTTLRR454 pKa = 11.84VFVDD458 pKa = 3.69DD459 pKa = 3.76YY460 pKa = 11.89GRR462 pKa = 11.84DD463 pKa = 3.49LFVSYY468 pKa = 9.96PVSHH472 pKa = 7.03TKK474 pKa = 9.95TDD476 pKa = 3.02ILLPDD481 pKa = 3.85EE482 pKa = 4.93ALHH485 pKa = 5.98PLEE488 pKa = 5.24PPVLLDD494 pKa = 4.66AYY496 pKa = 11.12SSDD499 pKa = 4.48YY500 pKa = 10.76YY501 pKa = 10.81LHH503 pKa = 6.96PSLWSRR509 pKa = 11.84RR510 pKa = 11.84KK511 pKa = 9.64RR512 pKa = 11.84KK513 pKa = 10.01RR514 pKa = 11.84SDD516 pKa = 2.5IYY518 pKa = 11.62NFFTDD523 pKa = 3.75GTVDD527 pKa = 3.15ATEE530 pKa = 3.84WW531 pKa = 3.26
MM1 pKa = 7.62EE2 pKa = 5.41PRR4 pKa = 11.84AKK6 pKa = 9.9RR7 pKa = 11.84QKK9 pKa = 10.17RR10 pKa = 11.84DD11 pKa = 3.45SVEE14 pKa = 3.53NLYY17 pKa = 10.18RR18 pKa = 11.84QCKK21 pKa = 8.4LTGQCPDD28 pKa = 3.27DD29 pKa = 4.54VINKK33 pKa = 9.42VEE35 pKa = 4.15GNTLADD41 pKa = 3.99RR42 pKa = 11.84LLKK45 pKa = 10.5ILGSVIYY52 pKa = 10.6LGGLGIGTGRR62 pKa = 11.84GSGGSTGYY70 pKa = 10.46RR71 pKa = 11.84PIGATTPRR79 pKa = 11.84VTDD82 pKa = 3.47STPIRR87 pKa = 11.84PTVPVDD93 pKa = 3.3PLGPTDD99 pKa = 4.24VLPVDD104 pKa = 3.63PAGPSIVPLTEE115 pKa = 3.78SGIPEE120 pKa = 3.91DD121 pKa = 4.43TIIEE125 pKa = 4.33TSGTAVTVNPTEE137 pKa = 4.19PPVITNIDD145 pKa = 4.31PISDD149 pKa = 3.57VTGVEE154 pKa = 4.11GQPTIITGEE163 pKa = 4.16DD164 pKa = 3.31DD165 pKa = 3.69SVAVLDD171 pKa = 4.36VQPSTPAPKK180 pKa = 9.96RR181 pKa = 11.84VTIGVRR187 pKa = 11.84PRR189 pKa = 11.84TSSPSEE195 pKa = 3.62PTLYY199 pKa = 10.2PIPEE203 pKa = 4.14PAAPDD208 pKa = 3.34FTVFVDD214 pKa = 4.21AQAVGEE220 pKa = 4.53TIGGEE225 pKa = 4.15YY226 pKa = 9.98IPLEE230 pKa = 4.4EE231 pKa = 5.01FSQIQNFEE239 pKa = 3.9IEE241 pKa = 4.32EE242 pKa = 4.05QQTLSSTPEE251 pKa = 3.53QRR253 pKa = 11.84LNRR256 pKa = 11.84AFQRR260 pKa = 11.84ARR262 pKa = 11.84DD263 pKa = 3.75LYY265 pKa = 10.82NRR267 pKa = 11.84QITQIRR273 pKa = 11.84TRR275 pKa = 11.84NLDD278 pKa = 3.42FLGAASRR285 pKa = 11.84AVQFEE290 pKa = 4.21FEE292 pKa = 4.35NPAFVDD298 pKa = 4.1DD299 pKa = 3.74VTLQFEE305 pKa = 4.18QDD307 pKa = 3.6VRR309 pKa = 11.84DD310 pKa = 3.84VAAAPDD316 pKa = 3.96PDD318 pKa = 3.57FRR320 pKa = 11.84DD321 pKa = 3.0IVTLHH326 pKa = 6.93RR327 pKa = 11.84ARR329 pKa = 11.84YY330 pKa = 7.11STTDD334 pKa = 2.98TGRR337 pKa = 11.84VRR339 pKa = 11.84VSRR342 pKa = 11.84LGKK345 pKa = 9.98RR346 pKa = 11.84GTIVTRR352 pKa = 11.84SGAQIGEE359 pKa = 4.3NVHH362 pKa = 6.74FYY364 pKa = 11.29YY365 pKa = 10.63DD366 pKa = 3.37VSTIGSDD373 pKa = 3.03AADD376 pKa = 4.67AIEE379 pKa = 4.85LSTIGQHH386 pKa = 6.19SGDD389 pKa = 3.93SAIVDD394 pKa = 3.93ALAEE398 pKa = 4.21SSFINPIEE406 pKa = 4.54NPDD409 pKa = 3.5TTYY412 pKa = 11.17TEE414 pKa = 4.76ADD416 pKa = 3.53LLDD419 pKa = 3.98TQVEE423 pKa = 4.44DD424 pKa = 4.06FDD426 pKa = 4.71GLHH429 pKa = 6.6LVLTSSGRR437 pKa = 11.84RR438 pKa = 11.84GDD440 pKa = 3.77VYY442 pKa = 10.94SIPTLPPGTTLRR454 pKa = 11.84VFVDD458 pKa = 3.69DD459 pKa = 3.76YY460 pKa = 11.89GRR462 pKa = 11.84DD463 pKa = 3.49LFVSYY468 pKa = 9.96PVSHH472 pKa = 7.03TKK474 pKa = 9.95TDD476 pKa = 3.02ILLPDD481 pKa = 3.85EE482 pKa = 4.93ALHH485 pKa = 5.98PLEE488 pKa = 5.24PPVLLDD494 pKa = 4.66AYY496 pKa = 11.12SSDD499 pKa = 4.48YY500 pKa = 10.76YY501 pKa = 10.81LHH503 pKa = 6.96PSLWSRR509 pKa = 11.84RR510 pKa = 11.84KK511 pKa = 9.64RR512 pKa = 11.84KK513 pKa = 10.01RR514 pKa = 11.84SDD516 pKa = 2.5IYY518 pKa = 11.62NFFTDD523 pKa = 3.75GTVDD527 pKa = 3.15ATEE530 pKa = 3.84WW531 pKa = 3.26
Molecular weight: 58.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2AM89|A0A2D2AM89_9PAPI Protein E7 OS=Gammapapillomavirus 24 OX=1961681 GN=E7 PE=3 SV=1
MM1 pKa = 7.53EE2 pKa = 4.87WLTDD6 pKa = 3.37RR7 pKa = 11.84FNAVQEE13 pKa = 4.27TLLNLIEE20 pKa = 5.37QGAQDD25 pKa = 4.68LDD27 pKa = 3.89SQIEE31 pKa = 4.04YY32 pKa = 9.14WNNVRR37 pKa = 11.84KK38 pKa = 10.07EE39 pKa = 4.14NVYY42 pKa = 9.03MYY44 pKa = 9.06YY45 pKa = 10.86AKK47 pKa = 10.48RR48 pKa = 11.84EE49 pKa = 4.13GLTHH53 pKa = 7.23IGLQPLPVPAVSEE66 pKa = 4.36YY67 pKa = 10.27KK68 pKa = 10.53AKK70 pKa = 10.38QAIHH74 pKa = 6.71LVLLLQSLKK83 pKa = 10.22KK84 pKa = 10.44SPYY87 pKa = 8.68ATEE90 pKa = 3.86SWTLQNTSEE99 pKa = 4.27EE100 pKa = 4.74LINTQPKK107 pKa = 10.61DD108 pKa = 3.61CFKK111 pKa = 10.38KK112 pKa = 9.74HH113 pKa = 6.45PYY115 pKa = 8.21TVEE118 pKa = 3.4VWFDD122 pKa = 3.18NDD124 pKa = 3.47RR125 pKa = 11.84NNSYY129 pKa = 10.75PYY131 pKa = 10.25INWDD135 pKa = 3.5AIYY138 pKa = 10.67YY139 pKa = 9.85QDD141 pKa = 5.31SQEE144 pKa = 4.16KK145 pKa = 6.18WHH147 pKa = 6.44KK148 pKa = 10.75VPGLVDD154 pKa = 3.71YY155 pKa = 11.46NGLYY159 pKa = 9.67YY160 pKa = 10.94NEE162 pKa = 4.3VGGDD166 pKa = 2.95RR167 pKa = 11.84VYY169 pKa = 10.36FTIFDD174 pKa = 3.9TDD176 pKa = 3.34AQKK179 pKa = 11.07YY180 pKa = 7.97GQTGMWTVHH189 pKa = 6.53FKK191 pKa = 10.91RR192 pKa = 11.84KK193 pKa = 7.85TLVAPISSSKK203 pKa = 10.37QSSAYY208 pKa = 10.02SGKK211 pKa = 8.23TPTSVSSSSKK221 pKa = 9.66DD222 pKa = 3.3TLPTKK227 pKa = 10.07EE228 pKa = 4.14SPRR231 pKa = 11.84RR232 pKa = 11.84LQRR235 pKa = 11.84PEE237 pKa = 4.02VGSSTGEE244 pKa = 3.83TTAVRR249 pKa = 11.84RR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84EE254 pKa = 3.73QGEE257 pKa = 4.26SSTDD261 pKa = 3.56GEE263 pKa = 4.42PTTTSKK269 pKa = 10.52RR270 pKa = 11.84RR271 pKa = 11.84RR272 pKa = 11.84GGGGGGGTGRR282 pKa = 11.84LGVSPEE288 pKa = 4.15EE289 pKa = 4.23VGSRR293 pKa = 11.84HH294 pKa = 5.55HH295 pKa = 6.53SATGRR300 pKa = 11.84HH301 pKa = 4.64RR302 pKa = 11.84TRR304 pKa = 11.84LEE306 pKa = 3.62QLKK309 pKa = 10.45EE310 pKa = 3.9EE311 pKa = 4.59ARR313 pKa = 11.84DD314 pKa = 3.53PPIIIVTGPANSLKK328 pKa = 9.67CWRR331 pKa = 11.84YY332 pKa = 9.64RR333 pKa = 11.84KK334 pKa = 9.51QNSNAFPFLAVSTIFTWVGGCGSAEE359 pKa = 4.0QARR362 pKa = 11.84MLIAFNTNEE371 pKa = 4.42EE372 pKa = 3.9RR373 pKa = 11.84TQFIKK378 pKa = 10.85YY379 pKa = 8.54VQFPKK384 pKa = 9.52QTTFALGQLDD394 pKa = 3.7KK395 pKa = 11.57LL396 pKa = 4.16
MM1 pKa = 7.53EE2 pKa = 4.87WLTDD6 pKa = 3.37RR7 pKa = 11.84FNAVQEE13 pKa = 4.27TLLNLIEE20 pKa = 5.37QGAQDD25 pKa = 4.68LDD27 pKa = 3.89SQIEE31 pKa = 4.04YY32 pKa = 9.14WNNVRR37 pKa = 11.84KK38 pKa = 10.07EE39 pKa = 4.14NVYY42 pKa = 9.03MYY44 pKa = 9.06YY45 pKa = 10.86AKK47 pKa = 10.48RR48 pKa = 11.84EE49 pKa = 4.13GLTHH53 pKa = 7.23IGLQPLPVPAVSEE66 pKa = 4.36YY67 pKa = 10.27KK68 pKa = 10.53AKK70 pKa = 10.38QAIHH74 pKa = 6.71LVLLLQSLKK83 pKa = 10.22KK84 pKa = 10.44SPYY87 pKa = 8.68ATEE90 pKa = 3.86SWTLQNTSEE99 pKa = 4.27EE100 pKa = 4.74LINTQPKK107 pKa = 10.61DD108 pKa = 3.61CFKK111 pKa = 10.38KK112 pKa = 9.74HH113 pKa = 6.45PYY115 pKa = 8.21TVEE118 pKa = 3.4VWFDD122 pKa = 3.18NDD124 pKa = 3.47RR125 pKa = 11.84NNSYY129 pKa = 10.75PYY131 pKa = 10.25INWDD135 pKa = 3.5AIYY138 pKa = 10.67YY139 pKa = 9.85QDD141 pKa = 5.31SQEE144 pKa = 4.16KK145 pKa = 6.18WHH147 pKa = 6.44KK148 pKa = 10.75VPGLVDD154 pKa = 3.71YY155 pKa = 11.46NGLYY159 pKa = 9.67YY160 pKa = 10.94NEE162 pKa = 4.3VGGDD166 pKa = 2.95RR167 pKa = 11.84VYY169 pKa = 10.36FTIFDD174 pKa = 3.9TDD176 pKa = 3.34AQKK179 pKa = 11.07YY180 pKa = 7.97GQTGMWTVHH189 pKa = 6.53FKK191 pKa = 10.91RR192 pKa = 11.84KK193 pKa = 7.85TLVAPISSSKK203 pKa = 10.37QSSAYY208 pKa = 10.02SGKK211 pKa = 8.23TPTSVSSSSKK221 pKa = 9.66DD222 pKa = 3.3TLPTKK227 pKa = 10.07EE228 pKa = 4.14SPRR231 pKa = 11.84RR232 pKa = 11.84LQRR235 pKa = 11.84PEE237 pKa = 4.02VGSSTGEE244 pKa = 3.83TTAVRR249 pKa = 11.84RR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84EE254 pKa = 3.73QGEE257 pKa = 4.26SSTDD261 pKa = 3.56GEE263 pKa = 4.42PTTTSKK269 pKa = 10.52RR270 pKa = 11.84RR271 pKa = 11.84RR272 pKa = 11.84GGGGGGGTGRR282 pKa = 11.84LGVSPEE288 pKa = 4.15EE289 pKa = 4.23VGSRR293 pKa = 11.84HH294 pKa = 5.55HH295 pKa = 6.53SATGRR300 pKa = 11.84HH301 pKa = 4.64RR302 pKa = 11.84TRR304 pKa = 11.84LEE306 pKa = 3.62QLKK309 pKa = 10.45EE310 pKa = 3.9EE311 pKa = 4.59ARR313 pKa = 11.84DD314 pKa = 3.53PPIIIVTGPANSLKK328 pKa = 9.67CWRR331 pKa = 11.84YY332 pKa = 9.64RR333 pKa = 11.84KK334 pKa = 9.51QNSNAFPFLAVSTIFTWVGGCGSAEE359 pKa = 4.0QARR362 pKa = 11.84MLIAFNTNEE371 pKa = 4.42EE372 pKa = 3.9RR373 pKa = 11.84TQFIKK378 pKa = 10.85YY379 pKa = 8.54VQFPKK384 pKa = 9.52QTTFALGQLDD394 pKa = 3.7KK395 pKa = 11.57LL396 pKa = 4.16
Molecular weight: 45.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2448 |
95 |
607 |
349.7 |
39.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.596 ± 0.495 | 2.206 ± 0.607 |
6.985 ± 0.748 | 6.25 ± 0.512 |
4.248 ± 0.415 | 5.76 ± 0.652 |
1.838 ± 0.227 | 5.27 ± 0.466 |
5.351 ± 0.905 | 8.864 ± 0.488 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.471 ± 0.322 | 4.453 ± 0.454 |
5.964 ± 1.017 | 5.351 ± 0.417 |
5.882 ± 0.692 | 6.618 ± 0.388 |
6.618 ± 0.761 | 6.168 ± 0.448 |
1.225 ± 0.284 | 3.881 ± 0.344 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
