
Sorex minutus polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
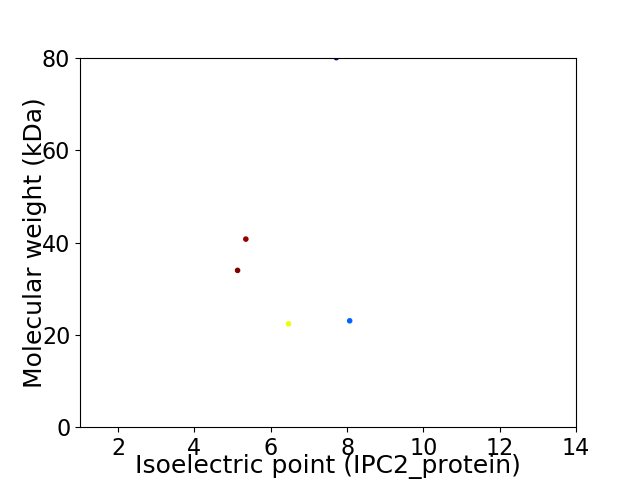
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A223PYN7|A0A223PYN7_9POLY small T antigen OS=Sorex minutus polyomavirus 1 OX=2560771 GN=STAg PE=4 SV=1
MM1 pKa = 7.44GGVISLAVEE10 pKa = 3.99LAGAISSLVAATEE23 pKa = 4.09LTAEE27 pKa = 4.97AILTGEE33 pKa = 4.12ALAAIEE39 pKa = 4.52SEE41 pKa = 4.52VASMVLVEE49 pKa = 5.18GFAEE53 pKa = 4.39ADD55 pKa = 3.12ALAALGLTAEE65 pKa = 4.47HH66 pKa = 6.58MSMLTAIPGMVEE78 pKa = 3.74QLASTGLLIQTLSGAAALFTAGIKK102 pKa = 10.43LSQHH106 pKa = 5.26EE107 pKa = 4.38VSLVNRR113 pKa = 11.84NTNMALQIWRR123 pKa = 11.84PDD125 pKa = 3.02LWEE128 pKa = 5.22LILPGFRR135 pKa = 11.84HH136 pKa = 6.01FEE138 pKa = 3.72WGFDD142 pKa = 3.55VLGDD146 pKa = 3.59WGHH149 pKa = 6.22SLVSAVGRR157 pKa = 11.84YY158 pKa = 6.27VWEE161 pKa = 3.97MLLTEE166 pKa = 4.01SRR168 pKa = 11.84RR169 pKa = 11.84RR170 pKa = 11.84LGMLGGNLIGEE181 pKa = 4.86GRR183 pKa = 11.84RR184 pKa = 11.84QAAEE188 pKa = 3.65QVAQLLEE195 pKa = 3.93GARR198 pKa = 11.84WVVTSPYY205 pKa = 10.12TGLQNYY211 pKa = 7.97YY212 pKa = 10.58RR213 pKa = 11.84SLPPMNVIQMRR224 pKa = 11.84QQARR228 pKa = 11.84LRR230 pKa = 11.84GQKK233 pKa = 10.47LIDD236 pKa = 3.54YY237 pKa = 9.8NRR239 pKa = 11.84VEE241 pKa = 4.63EE242 pKa = 4.39IDD244 pKa = 3.9PEE246 pKa = 4.32SGEE249 pKa = 4.59VIQKK253 pKa = 9.56YY254 pKa = 9.74GPPGGADD261 pKa = 3.2QPVTPDD267 pKa = 2.75WMLLMILGLYY277 pKa = 10.59GDD279 pKa = 4.27ITPLWGHH286 pKa = 5.71VLEE289 pKa = 4.71QEE291 pKa = 5.33HH292 pKa = 6.65ILSPKK297 pKa = 10.62NMIEE301 pKa = 4.11GPPAKK306 pKa = 9.77KK307 pKa = 9.58RR308 pKa = 11.84RR309 pKa = 11.84KK310 pKa = 8.08MM311 pKa = 3.39
MM1 pKa = 7.44GGVISLAVEE10 pKa = 3.99LAGAISSLVAATEE23 pKa = 4.09LTAEE27 pKa = 4.97AILTGEE33 pKa = 4.12ALAAIEE39 pKa = 4.52SEE41 pKa = 4.52VASMVLVEE49 pKa = 5.18GFAEE53 pKa = 4.39ADD55 pKa = 3.12ALAALGLTAEE65 pKa = 4.47HH66 pKa = 6.58MSMLTAIPGMVEE78 pKa = 3.74QLASTGLLIQTLSGAAALFTAGIKK102 pKa = 10.43LSQHH106 pKa = 5.26EE107 pKa = 4.38VSLVNRR113 pKa = 11.84NTNMALQIWRR123 pKa = 11.84PDD125 pKa = 3.02LWEE128 pKa = 5.22LILPGFRR135 pKa = 11.84HH136 pKa = 6.01FEE138 pKa = 3.72WGFDD142 pKa = 3.55VLGDD146 pKa = 3.59WGHH149 pKa = 6.22SLVSAVGRR157 pKa = 11.84YY158 pKa = 6.27VWEE161 pKa = 3.97MLLTEE166 pKa = 4.01SRR168 pKa = 11.84RR169 pKa = 11.84RR170 pKa = 11.84LGMLGGNLIGEE181 pKa = 4.86GRR183 pKa = 11.84RR184 pKa = 11.84QAAEE188 pKa = 3.65QVAQLLEE195 pKa = 3.93GARR198 pKa = 11.84WVVTSPYY205 pKa = 10.12TGLQNYY211 pKa = 7.97YY212 pKa = 10.58RR213 pKa = 11.84SLPPMNVIQMRR224 pKa = 11.84QQARR228 pKa = 11.84LRR230 pKa = 11.84GQKK233 pKa = 10.47LIDD236 pKa = 3.54YY237 pKa = 9.8NRR239 pKa = 11.84VEE241 pKa = 4.63EE242 pKa = 4.39IDD244 pKa = 3.9PEE246 pKa = 4.32SGEE249 pKa = 4.59VIQKK253 pKa = 9.56YY254 pKa = 9.74GPPGGADD261 pKa = 3.2QPVTPDD267 pKa = 2.75WMLLMILGLYY277 pKa = 10.59GDD279 pKa = 4.27ITPLWGHH286 pKa = 5.71VLEE289 pKa = 4.71QEE291 pKa = 5.33HH292 pKa = 6.65ILSPKK297 pKa = 10.62NMIEE301 pKa = 4.11GPPAKK306 pKa = 9.77KK307 pKa = 9.58RR308 pKa = 11.84RR309 pKa = 11.84KK310 pKa = 8.08MM311 pKa = 3.39
Molecular weight: 33.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A223PYN8|A0A223PYN8_9POLY Capsid protein VP1 OS=Sorex minutus polyomavirus 1 OX=2560771 GN=VP1 PE=3 SV=1
MM1 pKa = 7.84DD2 pKa = 6.58RR3 pKa = 11.84IFTSADD9 pKa = 3.53RR10 pKa = 11.84QLLISLLRR18 pKa = 11.84ISGDD22 pKa = 3.21TFGNVPAMARR32 pKa = 11.84AYY34 pKa = 9.7KK35 pKa = 10.24LAAKK39 pKa = 10.03RR40 pKa = 11.84LHH42 pKa = 6.69PDD44 pKa = 2.68KK45 pKa = 11.18GGNEE49 pKa = 4.08ADD51 pKa = 3.28MKK53 pKa = 10.91KK54 pKa = 10.73LNEE57 pKa = 4.01LWNKK61 pKa = 9.74FKK63 pKa = 11.4DD64 pKa = 3.4EE65 pKa = 4.2IYY67 pKa = 10.8RR68 pKa = 11.84MRR70 pKa = 11.84EE71 pKa = 3.58VKK73 pKa = 10.38PCVNPVITCIVLGTCSIFALIRR95 pKa = 11.84NTAQCMQNLLRR106 pKa = 11.84CCTCICCLLFKK117 pKa = 8.29QHH119 pKa = 6.96RR120 pKa = 11.84EE121 pKa = 3.63LKK123 pKa = 7.36ITYY126 pKa = 9.71RR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84CNVWGEE135 pKa = 4.32CYY137 pKa = 10.58CFFCYY142 pKa = 9.05YY143 pKa = 9.35TWFGVNCSLNAFTEE157 pKa = 4.07WLILLKK163 pKa = 10.81HH164 pKa = 6.36LDD166 pKa = 3.17WRR168 pKa = 11.84LLKK171 pKa = 10.39ISSAEE176 pKa = 3.78LDD178 pKa = 3.48SLGKK182 pKa = 10.61YY183 pKa = 9.62NFYY186 pKa = 10.8EE187 pKa = 3.63IFLIHH192 pKa = 6.62FVKK195 pKa = 10.81CC196 pKa = 3.56
MM1 pKa = 7.84DD2 pKa = 6.58RR3 pKa = 11.84IFTSADD9 pKa = 3.53RR10 pKa = 11.84QLLISLLRR18 pKa = 11.84ISGDD22 pKa = 3.21TFGNVPAMARR32 pKa = 11.84AYY34 pKa = 9.7KK35 pKa = 10.24LAAKK39 pKa = 10.03RR40 pKa = 11.84LHH42 pKa = 6.69PDD44 pKa = 2.68KK45 pKa = 11.18GGNEE49 pKa = 4.08ADD51 pKa = 3.28MKK53 pKa = 10.91KK54 pKa = 10.73LNEE57 pKa = 4.01LWNKK61 pKa = 9.74FKK63 pKa = 11.4DD64 pKa = 3.4EE65 pKa = 4.2IYY67 pKa = 10.8RR68 pKa = 11.84MRR70 pKa = 11.84EE71 pKa = 3.58VKK73 pKa = 10.38PCVNPVITCIVLGTCSIFALIRR95 pKa = 11.84NTAQCMQNLLRR106 pKa = 11.84CCTCICCLLFKK117 pKa = 8.29QHH119 pKa = 6.96RR120 pKa = 11.84EE121 pKa = 3.63LKK123 pKa = 7.36ITYY126 pKa = 9.71RR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84CNVWGEE135 pKa = 4.32CYY137 pKa = 10.58CFFCYY142 pKa = 9.05YY143 pKa = 9.35TWFGVNCSLNAFTEE157 pKa = 4.07WLILLKK163 pKa = 10.81HH164 pKa = 6.36LDD166 pKa = 3.17WRR168 pKa = 11.84LLKK171 pKa = 10.39ISSAEE176 pKa = 3.78LDD178 pKa = 3.48SLGKK182 pKa = 10.61YY183 pKa = 9.62NFYY186 pKa = 10.8EE187 pKa = 3.63IFLIHH192 pKa = 6.62FVKK195 pKa = 10.81CC196 pKa = 3.56
Molecular weight: 23.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1777 |
195 |
709 |
355.4 |
40.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.359 ± 0.974 | 2.251 ± 0.995 |
5.402 ± 0.773 | 5.909 ± 0.605 |
3.995 ± 1.074 | 7.034 ± 1.244 |
2.026 ± 0.38 | 5.684 ± 0.513 |
5.965 ± 1.381 | 10.242 ± 1.104 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.151 ± 0.599 | 4.671 ± 0.648 |
5.29 ± 0.729 | 3.995 ± 0.497 |
5.684 ± 0.437 | 6.584 ± 0.934 |
5.177 ± 0.645 | 5.684 ± 0.879 |
1.688 ± 0.55 | 3.208 ± 0.485 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
