
Mastomys coucha papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Pipapillomavirus; Pipapillomavirus 2
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
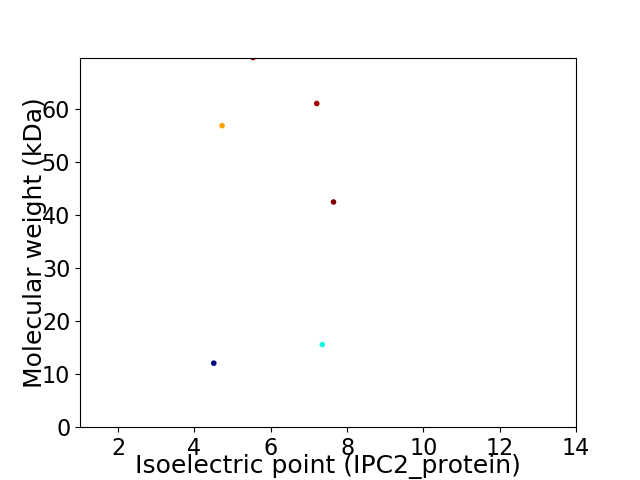
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|Q06RH5|Q06RH5_9PAPI Protein E6 OS=Mastomys coucha papillomavirus 2 OX=392505 GN=E6 PE=3 SV=1
MM1 pKa = 7.59LGPVPTLRR9 pKa = 11.84DD10 pKa = 3.26IEE12 pKa = 4.62LVDD15 pKa = 4.39LEE17 pKa = 4.38VLAAHH22 pKa = 6.61VPPADD27 pKa = 3.65QPDD30 pKa = 3.93VGSSSLSPDD39 pKa = 3.14SLGEE43 pKa = 4.03EE44 pKa = 4.48VEE46 pKa = 4.99LEE48 pKa = 3.84VDD50 pKa = 4.38PYY52 pKa = 11.03RR53 pKa = 11.84VATTCYY59 pKa = 10.5SCDD62 pKa = 3.16TVLRR66 pKa = 11.84FIVVAGCDD74 pKa = 3.38SLKK77 pKa = 10.81LFEE80 pKa = 4.95EE81 pKa = 4.83LLTEE85 pKa = 5.72DD86 pKa = 5.64FDD88 pKa = 4.95FLCPGCVKK96 pKa = 10.65DD97 pKa = 3.92RR98 pKa = 11.84LLRR101 pKa = 11.84KK102 pKa = 9.66RR103 pKa = 11.84NNGRR107 pKa = 11.84RR108 pKa = 11.84KK109 pKa = 9.84
MM1 pKa = 7.59LGPVPTLRR9 pKa = 11.84DD10 pKa = 3.26IEE12 pKa = 4.62LVDD15 pKa = 4.39LEE17 pKa = 4.38VLAAHH22 pKa = 6.61VPPADD27 pKa = 3.65QPDD30 pKa = 3.93VGSSSLSPDD39 pKa = 3.14SLGEE43 pKa = 4.03EE44 pKa = 4.48VEE46 pKa = 4.99LEE48 pKa = 3.84VDD50 pKa = 4.38PYY52 pKa = 11.03RR53 pKa = 11.84VATTCYY59 pKa = 10.5SCDD62 pKa = 3.16TVLRR66 pKa = 11.84FIVVAGCDD74 pKa = 3.38SLKK77 pKa = 10.81LFEE80 pKa = 4.95EE81 pKa = 4.83LLTEE85 pKa = 5.72DD86 pKa = 5.64FDD88 pKa = 4.95FLCPGCVKK96 pKa = 10.65DD97 pKa = 3.92RR98 pKa = 11.84LLRR101 pKa = 11.84KK102 pKa = 9.66RR103 pKa = 11.84NNGRR107 pKa = 11.84RR108 pKa = 11.84KK109 pKa = 9.84
Molecular weight: 12.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q06RH3|Q06RH3_9PAPI Replication protein E1 OS=Mastomys coucha papillomavirus 2 OX=392505 GN=E1 PE=3 SV=1
MM1 pKa = 7.48NSLEE5 pKa = 3.98ARR7 pKa = 11.84FDD9 pKa = 3.64AVQEE13 pKa = 4.32QILSLYY19 pKa = 10.1EE20 pKa = 4.07KK21 pKa = 10.71GSTDD25 pKa = 3.52LADD28 pKa = 3.39HH29 pKa = 5.68VKK31 pKa = 9.41YY32 pKa = 9.99WEE34 pKa = 4.7LIRR37 pKa = 11.84AEE39 pKa = 4.46GALEE43 pKa = 3.88YY44 pKa = 10.39CARR47 pKa = 11.84QSGRR51 pKa = 11.84NKK53 pKa = 10.44LGLHH57 pKa = 5.88SVPSSVGAEE66 pKa = 4.11SKK68 pKa = 10.61AKK70 pKa = 10.2KK71 pKa = 9.46AIHH74 pKa = 5.41MQMVLSSLQDD84 pKa = 3.13SSFGSEE90 pKa = 4.01PWTMADD96 pKa = 3.48TSLEE100 pKa = 4.13MYY102 pKa = 9.7EE103 pKa = 4.24RR104 pKa = 11.84TSPEE108 pKa = 3.58YY109 pKa = 9.76TFKK112 pKa = 11.12KK113 pKa = 9.88KK114 pKa = 10.61GSEE117 pKa = 4.11AEE119 pKa = 4.25VTFGSDD125 pKa = 3.12PDD127 pKa = 3.8NSVSYY132 pKa = 8.33MVWGCVYY139 pKa = 9.85TQDD142 pKa = 5.73EE143 pKa = 4.37DD144 pKa = 4.6GQWHH148 pKa = 5.74KK149 pKa = 10.81HH150 pKa = 4.5MSVVDD155 pKa = 3.84YY156 pKa = 11.22YY157 pKa = 10.96GVYY160 pKa = 9.96YY161 pKa = 10.45VDD163 pKa = 3.43HH164 pKa = 6.65SGVRR168 pKa = 11.84VYY170 pKa = 9.92YY171 pKa = 10.29HH172 pKa = 6.66QFDD175 pKa = 4.15EE176 pKa = 4.74DD177 pKa = 4.14SKK179 pKa = 10.16TFGGDD184 pKa = 2.79FWTVRR189 pKa = 11.84YY190 pKa = 9.62KK191 pKa = 11.04LQTFTSSPDD200 pKa = 3.09STTKK204 pKa = 10.65EE205 pKa = 3.96NEE207 pKa = 3.76QGLDD211 pKa = 3.67EE212 pKa = 4.9PCHH215 pKa = 5.69KK216 pKa = 10.12RR217 pKa = 11.84RR218 pKa = 11.84RR219 pKa = 11.84SSPCSTEE226 pKa = 4.05TIRR229 pKa = 11.84TPNRR233 pKa = 11.84RR234 pKa = 11.84ARR236 pKa = 11.84PSEE239 pKa = 3.92RR240 pKa = 11.84PQPTSGRR247 pKa = 11.84RR248 pKa = 11.84GRR250 pKa = 11.84LGQGEE255 pKa = 4.33RR256 pKa = 11.84TPEE259 pKa = 3.87GTRR262 pKa = 11.84PSPEE266 pKa = 3.94EE267 pKa = 3.3IGRR270 pKa = 11.84RR271 pKa = 11.84PEE273 pKa = 3.87GSEE276 pKa = 3.83RR277 pKa = 11.84SCQRR281 pKa = 11.84RR282 pKa = 11.84VPNPVHH288 pKa = 7.01CDD290 pKa = 3.36PPVILLKK297 pKa = 11.05GPTNTLKK304 pKa = 10.54CWRR307 pKa = 11.84NRR309 pKa = 11.84LRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84HH314 pKa = 5.6FRR316 pKa = 11.84PFIKK320 pKa = 10.15ISTAFSWVEE329 pKa = 3.74DD330 pKa = 3.9RR331 pKa = 11.84DD332 pKa = 4.14SGGVTNMLVAFLDD345 pKa = 3.71SQQRR349 pKa = 11.84DD350 pKa = 3.69VFLKK354 pKa = 9.34TVPIPKK360 pKa = 9.92GSVYY364 pKa = 10.98YY365 pKa = 10.48KK366 pKa = 11.24GNFEE370 pKa = 4.24GLL372 pKa = 3.46
MM1 pKa = 7.48NSLEE5 pKa = 3.98ARR7 pKa = 11.84FDD9 pKa = 3.64AVQEE13 pKa = 4.32QILSLYY19 pKa = 10.1EE20 pKa = 4.07KK21 pKa = 10.71GSTDD25 pKa = 3.52LADD28 pKa = 3.39HH29 pKa = 5.68VKK31 pKa = 9.41YY32 pKa = 9.99WEE34 pKa = 4.7LIRR37 pKa = 11.84AEE39 pKa = 4.46GALEE43 pKa = 3.88YY44 pKa = 10.39CARR47 pKa = 11.84QSGRR51 pKa = 11.84NKK53 pKa = 10.44LGLHH57 pKa = 5.88SVPSSVGAEE66 pKa = 4.11SKK68 pKa = 10.61AKK70 pKa = 10.2KK71 pKa = 9.46AIHH74 pKa = 5.41MQMVLSSLQDD84 pKa = 3.13SSFGSEE90 pKa = 4.01PWTMADD96 pKa = 3.48TSLEE100 pKa = 4.13MYY102 pKa = 9.7EE103 pKa = 4.24RR104 pKa = 11.84TSPEE108 pKa = 3.58YY109 pKa = 9.76TFKK112 pKa = 11.12KK113 pKa = 9.88KK114 pKa = 10.61GSEE117 pKa = 4.11AEE119 pKa = 4.25VTFGSDD125 pKa = 3.12PDD127 pKa = 3.8NSVSYY132 pKa = 8.33MVWGCVYY139 pKa = 9.85TQDD142 pKa = 5.73EE143 pKa = 4.37DD144 pKa = 4.6GQWHH148 pKa = 5.74KK149 pKa = 10.81HH150 pKa = 4.5MSVVDD155 pKa = 3.84YY156 pKa = 11.22YY157 pKa = 10.96GVYY160 pKa = 9.96YY161 pKa = 10.45VDD163 pKa = 3.43HH164 pKa = 6.65SGVRR168 pKa = 11.84VYY170 pKa = 9.92YY171 pKa = 10.29HH172 pKa = 6.66QFDD175 pKa = 4.15EE176 pKa = 4.74DD177 pKa = 4.14SKK179 pKa = 10.16TFGGDD184 pKa = 2.79FWTVRR189 pKa = 11.84YY190 pKa = 9.62KK191 pKa = 11.04LQTFTSSPDD200 pKa = 3.09STTKK204 pKa = 10.65EE205 pKa = 3.96NEE207 pKa = 3.76QGLDD211 pKa = 3.67EE212 pKa = 4.9PCHH215 pKa = 5.69KK216 pKa = 10.12RR217 pKa = 11.84RR218 pKa = 11.84RR219 pKa = 11.84SSPCSTEE226 pKa = 4.05TIRR229 pKa = 11.84TPNRR233 pKa = 11.84RR234 pKa = 11.84ARR236 pKa = 11.84PSEE239 pKa = 3.92RR240 pKa = 11.84PQPTSGRR247 pKa = 11.84RR248 pKa = 11.84GRR250 pKa = 11.84LGQGEE255 pKa = 4.33RR256 pKa = 11.84TPEE259 pKa = 3.87GTRR262 pKa = 11.84PSPEE266 pKa = 3.94EE267 pKa = 3.3IGRR270 pKa = 11.84RR271 pKa = 11.84PEE273 pKa = 3.87GSEE276 pKa = 3.83RR277 pKa = 11.84SCQRR281 pKa = 11.84RR282 pKa = 11.84VPNPVHH288 pKa = 7.01CDD290 pKa = 3.36PPVILLKK297 pKa = 11.05GPTNTLKK304 pKa = 10.54CWRR307 pKa = 11.84NRR309 pKa = 11.84LRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84HH314 pKa = 5.6FRR316 pKa = 11.84PFIKK320 pKa = 10.15ISTAFSWVEE329 pKa = 3.74DD330 pKa = 3.9RR331 pKa = 11.84DD332 pKa = 4.14SGGVTNMLVAFLDD345 pKa = 3.71SQQRR349 pKa = 11.84DD350 pKa = 3.69VFLKK354 pKa = 9.34TVPIPKK360 pKa = 9.92GSVYY364 pKa = 10.98YY365 pKa = 10.48KK366 pKa = 11.24GNFEE370 pKa = 4.24GLL372 pKa = 3.46
Molecular weight: 42.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2306 |
109 |
614 |
384.3 |
42.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.637 ± 0.849 | 2.298 ± 0.576 |
6.548 ± 0.323 | 6.071 ± 0.588 |
4.944 ± 0.345 | 7.329 ± 1.077 |
1.951 ± 0.249 | 3.903 ± 0.853 |
4.337 ± 0.875 | 8.066 ± 0.808 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.255 ± 0.417 | 4.163 ± 0.649 |
6.288 ± 0.96 | 3.99 ± 0.483 |
6.852 ± 0.437 | 7.502 ± 0.661 |
6.808 ± 0.4 | 7.112 ± 0.772 |
1.171 ± 0.311 | 2.775 ± 0.361 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
