
Sphingomonas changbaiensis NBRC 104936
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas; Sphingomonas changbaiensis
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
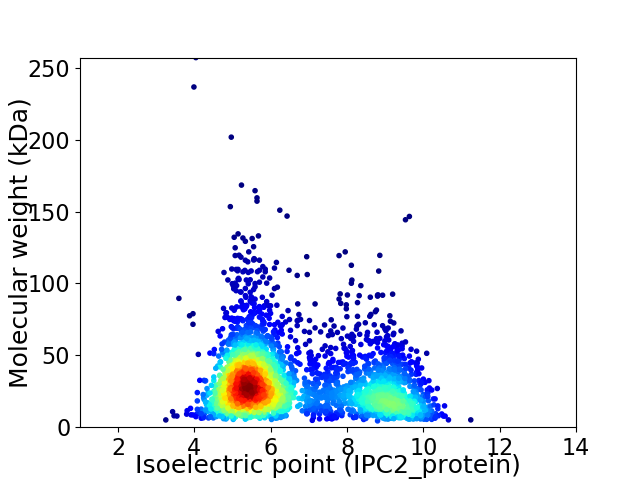
Virtual 2D-PAGE plot for 3151 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E9MNF1|A0A0E9MNF1_9SPHN TPR_REGION domain-containing protein OS=Sphingomonas changbaiensis NBRC 104936 OX=1219043 GN=SCH01S_17_00140 PE=4 SV=1
MM1 pKa = 7.63TIFNVSNSTQLQSALGSATGGDD23 pKa = 4.23TIVLADD29 pKa = 3.7GDD31 pKa = 4.16YY32 pKa = 11.57GKK34 pKa = 11.26VNVYY38 pKa = 8.18TRR40 pKa = 11.84NYY42 pKa = 9.4ASTVTIEE49 pKa = 3.62AATPGGAHH57 pKa = 7.37LDD59 pKa = 3.69GLFVANATNISFAGLDD75 pKa = 3.24VGRR78 pKa = 11.84ALGATEE84 pKa = 4.3PDD86 pKa = 3.7YY87 pKa = 10.91TQLSSVSNSSNITMSHH103 pKa = 4.84ITVHH107 pKa = 6.58GSLDD111 pKa = 3.63NDD113 pKa = 4.11PANDD117 pKa = 3.63GMGMMVRR124 pKa = 11.84NVAGFSISDD133 pKa = 3.52STFGDD138 pKa = 4.4LYY140 pKa = 10.94RR141 pKa = 11.84GIVLQNNTNATVQNNTIKK159 pKa = 10.11TIRR162 pKa = 11.84SDD164 pKa = 4.15GIISIANDD172 pKa = 4.96GITIAGNHH180 pKa = 6.24IGEE183 pKa = 4.45FHH185 pKa = 7.16PKK187 pKa = 10.47LGDD190 pKa = 3.46HH191 pKa = 7.31ADD193 pKa = 5.39AIQFWNTGQTKK204 pKa = 9.15GQTDD208 pKa = 3.44ITVKK212 pKa = 11.02DD213 pKa = 3.52NVIYY217 pKa = 10.47QSYY220 pKa = 10.35FSGIDD225 pKa = 3.11QTGVQGIFMSDD236 pKa = 3.28PLSYY240 pKa = 10.48GYY242 pKa = 11.18KK243 pKa = 10.25NVLIQNNLLYY253 pKa = 11.27SNDD256 pKa = 3.37AYY258 pKa = 11.5NGIFVNGATGAQIIGNTVLSHH279 pKa = 6.88SSDD282 pKa = 3.44SKK284 pKa = 11.31LFWIDD289 pKa = 3.34VLNSDD294 pKa = 5.09QVSLQDD300 pKa = 3.63NLTDD304 pKa = 5.79RR305 pKa = 11.84IITSNVTNFAQSGNVDD321 pKa = 4.02FSVNAGARR329 pKa = 11.84AMLPNLANPAGVEE342 pKa = 4.08DD343 pKa = 4.71LVASGVGYY351 pKa = 10.03HH352 pKa = 6.84LPPPPTPITPPAPLPIDD369 pKa = 3.68FTGDD373 pKa = 3.39DD374 pKa = 4.24ANNLINANAADD385 pKa = 3.79NNIDD389 pKa = 3.63GGKK392 pKa = 10.21GADD395 pKa = 3.54AMYY398 pKa = 11.0GLAGNDD404 pKa = 3.4HH405 pKa = 6.92YY406 pKa = 11.38IVDD409 pKa = 3.92NAGDD413 pKa = 3.79RR414 pKa = 11.84VFEE417 pKa = 4.51DD418 pKa = 3.76ANAGHH423 pKa = 7.05DD424 pKa = 3.85VVSTSVTYY432 pKa = 10.62SLQAGTSVEE441 pKa = 4.12VLQVLGPDD449 pKa = 3.07TTTAINLTGNEE460 pKa = 4.33LANTLIGNAGKK471 pKa = 10.35NILDD475 pKa = 4.04GGAGADD481 pKa = 4.1EE482 pKa = 4.19LHH484 pKa = 6.62GLAGDD489 pKa = 3.43DD490 pKa = 4.82CYY492 pKa = 11.03FVDD495 pKa = 3.71HH496 pKa = 7.09AGDD499 pKa = 3.35HH500 pKa = 5.97VYY502 pKa = 10.53EE503 pKa = 4.42VAGEE507 pKa = 4.56GYY509 pKa = 8.41DD510 pKa = 3.73TVCASVSYY518 pKa = 10.63ALQAGTSIEE527 pKa = 4.08VLQTNDD533 pKa = 2.98VAGTAAINLTGNEE546 pKa = 4.06LVNTVFGNGGANILDD561 pKa = 4.13GGAGADD567 pKa = 3.49KK568 pKa = 10.91LQGFGGDD575 pKa = 3.62DD576 pKa = 3.52SYY578 pKa = 12.11LVDD581 pKa = 3.51NARR584 pKa = 11.84DD585 pKa = 3.75SVYY588 pKa = 10.37EE589 pKa = 3.8AAGGGRR595 pKa = 11.84DD596 pKa = 3.44TVSASVSYY604 pKa = 9.04TLQPGTYY611 pKa = 9.9VEE613 pKa = 4.3VLQTNDD619 pKa = 3.56DD620 pKa = 4.09GGTATLDD627 pKa = 3.38LSGNDD632 pKa = 3.23FANVVRR638 pKa = 11.84GNAGNNTLQGRR649 pKa = 11.84GGDD652 pKa = 3.74DD653 pKa = 3.3VLIGLGGDD661 pKa = 3.33DD662 pKa = 4.28RR663 pKa = 11.84LYY665 pKa = 11.19GQAGNDD671 pKa = 3.36TFVFAPGFGRR681 pKa = 11.84DD682 pKa = 3.5MVGDD686 pKa = 4.57FAAHH690 pKa = 5.97SAGGGDD696 pKa = 3.7LVAFQGKK703 pKa = 9.5AFSGFDD709 pKa = 3.59DD710 pKa = 4.49LMAHH714 pKa = 7.46AAQKK718 pKa = 9.17GTDD721 pKa = 3.38VVFTLDD727 pKa = 3.57ANTSLTLHH735 pKa = 5.96NVQLAALSAQDD746 pKa = 3.32FTFAA750 pKa = 4.84
MM1 pKa = 7.63TIFNVSNSTQLQSALGSATGGDD23 pKa = 4.23TIVLADD29 pKa = 3.7GDD31 pKa = 4.16YY32 pKa = 11.57GKK34 pKa = 11.26VNVYY38 pKa = 8.18TRR40 pKa = 11.84NYY42 pKa = 9.4ASTVTIEE49 pKa = 3.62AATPGGAHH57 pKa = 7.37LDD59 pKa = 3.69GLFVANATNISFAGLDD75 pKa = 3.24VGRR78 pKa = 11.84ALGATEE84 pKa = 4.3PDD86 pKa = 3.7YY87 pKa = 10.91TQLSSVSNSSNITMSHH103 pKa = 4.84ITVHH107 pKa = 6.58GSLDD111 pKa = 3.63NDD113 pKa = 4.11PANDD117 pKa = 3.63GMGMMVRR124 pKa = 11.84NVAGFSISDD133 pKa = 3.52STFGDD138 pKa = 4.4LYY140 pKa = 10.94RR141 pKa = 11.84GIVLQNNTNATVQNNTIKK159 pKa = 10.11TIRR162 pKa = 11.84SDD164 pKa = 4.15GIISIANDD172 pKa = 4.96GITIAGNHH180 pKa = 6.24IGEE183 pKa = 4.45FHH185 pKa = 7.16PKK187 pKa = 10.47LGDD190 pKa = 3.46HH191 pKa = 7.31ADD193 pKa = 5.39AIQFWNTGQTKK204 pKa = 9.15GQTDD208 pKa = 3.44ITVKK212 pKa = 11.02DD213 pKa = 3.52NVIYY217 pKa = 10.47QSYY220 pKa = 10.35FSGIDD225 pKa = 3.11QTGVQGIFMSDD236 pKa = 3.28PLSYY240 pKa = 10.48GYY242 pKa = 11.18KK243 pKa = 10.25NVLIQNNLLYY253 pKa = 11.27SNDD256 pKa = 3.37AYY258 pKa = 11.5NGIFVNGATGAQIIGNTVLSHH279 pKa = 6.88SSDD282 pKa = 3.44SKK284 pKa = 11.31LFWIDD289 pKa = 3.34VLNSDD294 pKa = 5.09QVSLQDD300 pKa = 3.63NLTDD304 pKa = 5.79RR305 pKa = 11.84IITSNVTNFAQSGNVDD321 pKa = 4.02FSVNAGARR329 pKa = 11.84AMLPNLANPAGVEE342 pKa = 4.08DD343 pKa = 4.71LVASGVGYY351 pKa = 10.03HH352 pKa = 6.84LPPPPTPITPPAPLPIDD369 pKa = 3.68FTGDD373 pKa = 3.39DD374 pKa = 4.24ANNLINANAADD385 pKa = 3.79NNIDD389 pKa = 3.63GGKK392 pKa = 10.21GADD395 pKa = 3.54AMYY398 pKa = 11.0GLAGNDD404 pKa = 3.4HH405 pKa = 6.92YY406 pKa = 11.38IVDD409 pKa = 3.92NAGDD413 pKa = 3.79RR414 pKa = 11.84VFEE417 pKa = 4.51DD418 pKa = 3.76ANAGHH423 pKa = 7.05DD424 pKa = 3.85VVSTSVTYY432 pKa = 10.62SLQAGTSVEE441 pKa = 4.12VLQVLGPDD449 pKa = 3.07TTTAINLTGNEE460 pKa = 4.33LANTLIGNAGKK471 pKa = 10.35NILDD475 pKa = 4.04GGAGADD481 pKa = 4.1EE482 pKa = 4.19LHH484 pKa = 6.62GLAGDD489 pKa = 3.43DD490 pKa = 4.82CYY492 pKa = 11.03FVDD495 pKa = 3.71HH496 pKa = 7.09AGDD499 pKa = 3.35HH500 pKa = 5.97VYY502 pKa = 10.53EE503 pKa = 4.42VAGEE507 pKa = 4.56GYY509 pKa = 8.41DD510 pKa = 3.73TVCASVSYY518 pKa = 10.63ALQAGTSIEE527 pKa = 4.08VLQTNDD533 pKa = 2.98VAGTAAINLTGNEE546 pKa = 4.06LVNTVFGNGGANILDD561 pKa = 4.13GGAGADD567 pKa = 3.49KK568 pKa = 10.91LQGFGGDD575 pKa = 3.62DD576 pKa = 3.52SYY578 pKa = 12.11LVDD581 pKa = 3.51NARR584 pKa = 11.84DD585 pKa = 3.75SVYY588 pKa = 10.37EE589 pKa = 3.8AAGGGRR595 pKa = 11.84DD596 pKa = 3.44TVSASVSYY604 pKa = 9.04TLQPGTYY611 pKa = 9.9VEE613 pKa = 4.3VLQTNDD619 pKa = 3.56DD620 pKa = 4.09GGTATLDD627 pKa = 3.38LSGNDD632 pKa = 3.23FANVVRR638 pKa = 11.84GNAGNNTLQGRR649 pKa = 11.84GGDD652 pKa = 3.74DD653 pKa = 3.3VLIGLGGDD661 pKa = 3.33DD662 pKa = 4.28RR663 pKa = 11.84LYY665 pKa = 11.19GQAGNDD671 pKa = 3.36TFVFAPGFGRR681 pKa = 11.84DD682 pKa = 3.5MVGDD686 pKa = 4.57FAAHH690 pKa = 5.97SAGGGDD696 pKa = 3.7LVAFQGKK703 pKa = 9.5AFSGFDD709 pKa = 3.59DD710 pKa = 4.49LMAHH714 pKa = 7.46AAQKK718 pKa = 9.17GTDD721 pKa = 3.38VVFTLDD727 pKa = 3.57ANTSLTLHH735 pKa = 5.96NVQLAALSAQDD746 pKa = 3.32FTFAA750 pKa = 4.84
Molecular weight: 77.5 kDa
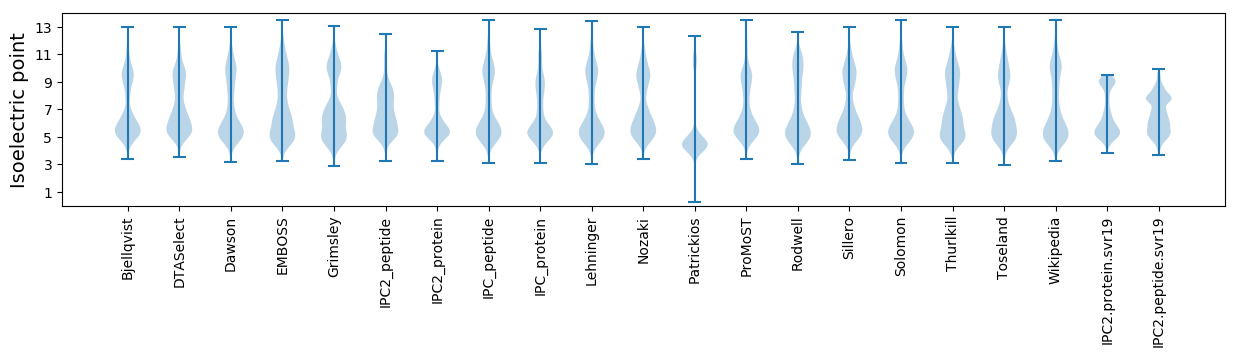
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E9MP55|A0A0E9MP55_9SPHN Uncharacterized protein OS=Sphingomonas changbaiensis NBRC 104936 OX=1219043 GN=SCH01S_28_00750 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.32VIAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.89VLSAA44 pKa = 4.11
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.32VIAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.89VLSAA44 pKa = 4.11
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
976675 |
41 |
2672 |
310.0 |
33.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.403 ± 0.066 | 0.75 ± 0.013 |
5.932 ± 0.034 | 5.51 ± 0.042 |
3.555 ± 0.03 | 8.95 ± 0.052 |
1.972 ± 0.02 | 4.858 ± 0.026 |
3.011 ± 0.035 | 10.0 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.262 ± 0.022 | 2.504 ± 0.033 |
5.541 ± 0.034 | 3.102 ± 0.027 |
7.597 ± 0.052 | 5.084 ± 0.036 |
5.163 ± 0.033 | 7.197 ± 0.036 |
1.431 ± 0.021 | 2.18 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
