
Carrot Ch virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Trivirinae; Chordovirus
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
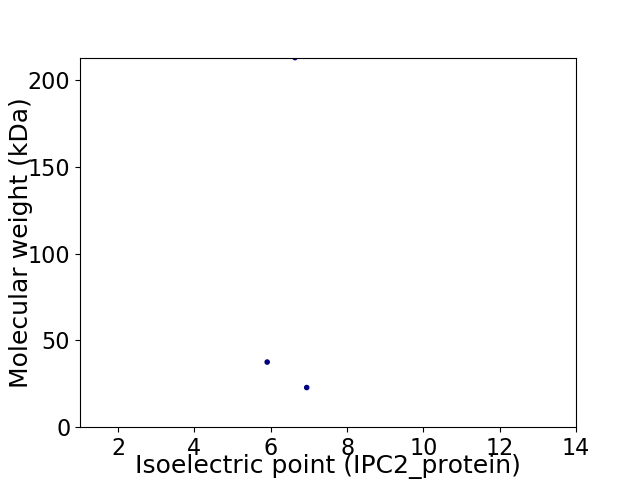
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0A0P9V6|A0A0A0P9V6_9VIRU ORF1 OS=Carrot Ch virus 2 OX=1425362 PE=4 SV=1
MM1 pKa = 7.35SLVDD5 pKa = 3.77INKK8 pKa = 9.24FCALVSKK15 pKa = 10.39GASSVEE21 pKa = 4.33AIDD24 pKa = 4.08KK25 pKa = 10.95GNIYY29 pKa = 10.66GDD31 pKa = 3.94SSWLKK36 pKa = 9.7FNQVSAIRR44 pKa = 11.84KK45 pKa = 8.08FEE47 pKa = 4.61SNISIEE53 pKa = 4.71LDD55 pKa = 3.78DD56 pKa = 4.63GDD58 pKa = 4.24KK59 pKa = 11.33SLVIPNIPLVSSFEE73 pKa = 4.1VATVKK78 pKa = 10.68KK79 pKa = 10.27SLKK82 pKa = 9.69NANFLHH88 pKa = 6.65LGGIVISIQALFAEE102 pKa = 4.75NKK104 pKa = 9.65GVTGTAVLADD114 pKa = 3.87KK115 pKa = 10.53RR116 pKa = 11.84WNNLDD121 pKa = 3.25QAIIGAFHH129 pKa = 6.98FNLDD133 pKa = 3.03KK134 pKa = 11.03RR135 pKa = 11.84RR136 pKa = 11.84ADD138 pKa = 4.21FIMRR142 pKa = 11.84PNFDD146 pKa = 3.06VSLNDD151 pKa = 4.19PMLCDD156 pKa = 5.27SISLMLKK163 pKa = 10.37FEE165 pKa = 4.39NLDD168 pKa = 3.6MQVSSIPINVSVGLIARR185 pKa = 11.84FYY187 pKa = 9.67NTIDD191 pKa = 3.56PGIKK195 pKa = 9.96FADD198 pKa = 4.41EE199 pKa = 3.95EE200 pKa = 4.7LKK202 pKa = 9.78MQGLIEE208 pKa = 4.94AEE210 pKa = 4.65GIDD213 pKa = 4.4KK214 pKa = 11.11NDD216 pKa = 3.64MLHH219 pKa = 7.18SIGDD223 pKa = 3.83LADD226 pKa = 3.68TFSNPAIFNPTIVNNRR242 pKa = 11.84KK243 pKa = 9.03YY244 pKa = 10.98DD245 pKa = 3.44RR246 pKa = 11.84GLFKK250 pKa = 11.05DD251 pKa = 3.55KK252 pKa = 11.35GFIKK256 pKa = 10.19QIRR259 pKa = 11.84GSRR262 pKa = 11.84SDD264 pKa = 3.25DD265 pKa = 3.37SKK267 pKa = 11.22ALRR270 pKa = 11.84RR271 pKa = 11.84VGRR274 pKa = 11.84FNAPSSSRR282 pKa = 11.84RR283 pKa = 11.84FDD285 pKa = 3.58QIKK288 pKa = 10.26VDD290 pKa = 3.59PLFYY294 pKa = 11.04DD295 pKa = 3.79KK296 pKa = 10.72IKK298 pKa = 10.68RR299 pKa = 11.84GNINSWLNNEE309 pKa = 4.4LPKK312 pKa = 10.61SSEE315 pKa = 4.13RR316 pKa = 11.84EE317 pKa = 3.67EE318 pKa = 3.9EE319 pKa = 4.4RR320 pKa = 11.84YY321 pKa = 10.19SEE323 pKa = 4.18TGSEE327 pKa = 4.03GRR329 pKa = 11.84VEE331 pKa = 4.21IVEE334 pKa = 4.28HH335 pKa = 6.26
MM1 pKa = 7.35SLVDD5 pKa = 3.77INKK8 pKa = 9.24FCALVSKK15 pKa = 10.39GASSVEE21 pKa = 4.33AIDD24 pKa = 4.08KK25 pKa = 10.95GNIYY29 pKa = 10.66GDD31 pKa = 3.94SSWLKK36 pKa = 9.7FNQVSAIRR44 pKa = 11.84KK45 pKa = 8.08FEE47 pKa = 4.61SNISIEE53 pKa = 4.71LDD55 pKa = 3.78DD56 pKa = 4.63GDD58 pKa = 4.24KK59 pKa = 11.33SLVIPNIPLVSSFEE73 pKa = 4.1VATVKK78 pKa = 10.68KK79 pKa = 10.27SLKK82 pKa = 9.69NANFLHH88 pKa = 6.65LGGIVISIQALFAEE102 pKa = 4.75NKK104 pKa = 9.65GVTGTAVLADD114 pKa = 3.87KK115 pKa = 10.53RR116 pKa = 11.84WNNLDD121 pKa = 3.25QAIIGAFHH129 pKa = 6.98FNLDD133 pKa = 3.03KK134 pKa = 11.03RR135 pKa = 11.84RR136 pKa = 11.84ADD138 pKa = 4.21FIMRR142 pKa = 11.84PNFDD146 pKa = 3.06VSLNDD151 pKa = 4.19PMLCDD156 pKa = 5.27SISLMLKK163 pKa = 10.37FEE165 pKa = 4.39NLDD168 pKa = 3.6MQVSSIPINVSVGLIARR185 pKa = 11.84FYY187 pKa = 9.67NTIDD191 pKa = 3.56PGIKK195 pKa = 9.96FADD198 pKa = 4.41EE199 pKa = 3.95EE200 pKa = 4.7LKK202 pKa = 9.78MQGLIEE208 pKa = 4.94AEE210 pKa = 4.65GIDD213 pKa = 4.4KK214 pKa = 11.11NDD216 pKa = 3.64MLHH219 pKa = 7.18SIGDD223 pKa = 3.83LADD226 pKa = 3.68TFSNPAIFNPTIVNNRR242 pKa = 11.84KK243 pKa = 9.03YY244 pKa = 10.98DD245 pKa = 3.44RR246 pKa = 11.84GLFKK250 pKa = 11.05DD251 pKa = 3.55KK252 pKa = 11.35GFIKK256 pKa = 10.19QIRR259 pKa = 11.84GSRR262 pKa = 11.84SDD264 pKa = 3.25DD265 pKa = 3.37SKK267 pKa = 11.22ALRR270 pKa = 11.84RR271 pKa = 11.84VGRR274 pKa = 11.84FNAPSSSRR282 pKa = 11.84RR283 pKa = 11.84FDD285 pKa = 3.58QIKK288 pKa = 10.26VDD290 pKa = 3.59PLFYY294 pKa = 11.04DD295 pKa = 3.79KK296 pKa = 10.72IKK298 pKa = 10.68RR299 pKa = 11.84GNINSWLNNEE309 pKa = 4.4LPKK312 pKa = 10.61SSEE315 pKa = 4.13RR316 pKa = 11.84EE317 pKa = 3.67EE318 pKa = 3.9EE319 pKa = 4.4RR320 pKa = 11.84YY321 pKa = 10.19SEE323 pKa = 4.18TGSEE327 pKa = 4.03GRR329 pKa = 11.84VEE331 pKa = 4.21IVEE334 pKa = 4.28HH335 pKa = 6.26
Molecular weight: 37.53 kDa
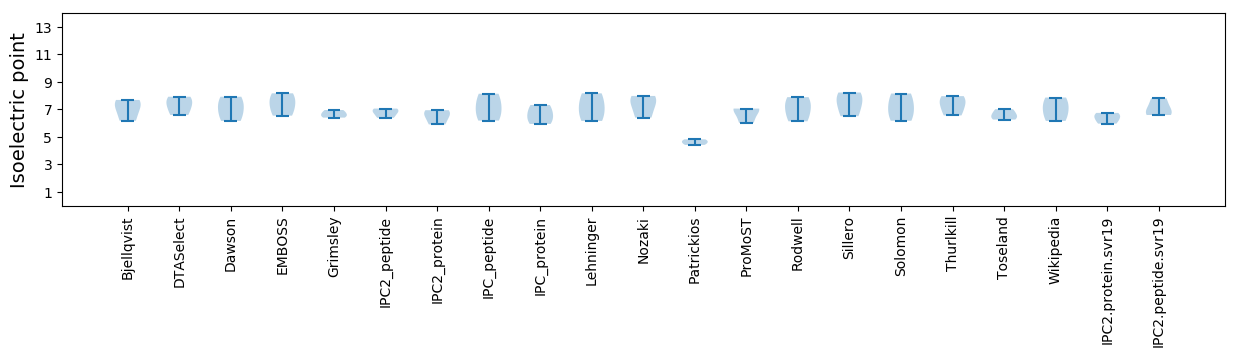
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0P507|A0A0A0P507_9VIRU ORF2 OS=Carrot Ch virus 2 OX=1425362 PE=4 SV=1
MM1 pKa = 7.8SYY3 pKa = 11.19LSLQRR8 pKa = 11.84EE9 pKa = 3.98RR10 pKa = 11.84RR11 pKa = 11.84SDD13 pKa = 3.37IVKK16 pKa = 10.28LALKK20 pKa = 10.57EE21 pKa = 3.95EE22 pKa = 4.16LRR24 pKa = 11.84SLNTEE29 pKa = 3.35IDD31 pKa = 3.42EE32 pKa = 4.47KK33 pKa = 10.8GTYY36 pKa = 9.2FKK38 pKa = 10.79QVLEE42 pKa = 4.09YY43 pKa = 10.57YY44 pKa = 10.25IMYY47 pKa = 10.08FVLNMAEE54 pKa = 4.54MGTSPDD60 pKa = 3.04TVYY63 pKa = 10.66PSEE66 pKa = 4.09NFEE69 pKa = 4.56LKK71 pKa = 10.27PVTMKK76 pKa = 10.84SGDD79 pKa = 3.6EE80 pKa = 3.94PKK82 pKa = 10.64TFILKK87 pKa = 9.63MNLITCIDD95 pKa = 4.76RR96 pKa = 11.84IQDD99 pKa = 3.52LSEE102 pKa = 3.86NRR104 pKa = 11.84VDD106 pKa = 6.05LGDD109 pKa = 3.48LTLRR113 pKa = 11.84SFMRR117 pKa = 11.84PFADD121 pKa = 2.98IAKK124 pKa = 9.67KK125 pKa = 9.89QLQKK129 pKa = 10.8RR130 pKa = 11.84GVDD133 pKa = 4.0GILARR138 pKa = 11.84KK139 pKa = 8.58YY140 pKa = 10.27PRR142 pKa = 11.84VCRR145 pKa = 11.84GAKK148 pKa = 10.19HH149 pKa = 5.88MGFDD153 pKa = 4.74FNQDD157 pKa = 3.43LQSSEE162 pKa = 4.11LSNSEE167 pKa = 3.64MRR169 pKa = 11.84VKK171 pKa = 10.72QNLGRR176 pKa = 11.84VMFNRR181 pKa = 11.84GLKK184 pKa = 9.07VANLEE189 pKa = 4.18EE190 pKa = 4.39SSSIDD195 pKa = 3.54FEE197 pKa = 4.47
MM1 pKa = 7.8SYY3 pKa = 11.19LSLQRR8 pKa = 11.84EE9 pKa = 3.98RR10 pKa = 11.84RR11 pKa = 11.84SDD13 pKa = 3.37IVKK16 pKa = 10.28LALKK20 pKa = 10.57EE21 pKa = 3.95EE22 pKa = 4.16LRR24 pKa = 11.84SLNTEE29 pKa = 3.35IDD31 pKa = 3.42EE32 pKa = 4.47KK33 pKa = 10.8GTYY36 pKa = 9.2FKK38 pKa = 10.79QVLEE42 pKa = 4.09YY43 pKa = 10.57YY44 pKa = 10.25IMYY47 pKa = 10.08FVLNMAEE54 pKa = 4.54MGTSPDD60 pKa = 3.04TVYY63 pKa = 10.66PSEE66 pKa = 4.09NFEE69 pKa = 4.56LKK71 pKa = 10.27PVTMKK76 pKa = 10.84SGDD79 pKa = 3.6EE80 pKa = 3.94PKK82 pKa = 10.64TFILKK87 pKa = 9.63MNLITCIDD95 pKa = 4.76RR96 pKa = 11.84IQDD99 pKa = 3.52LSEE102 pKa = 3.86NRR104 pKa = 11.84VDD106 pKa = 6.05LGDD109 pKa = 3.48LTLRR113 pKa = 11.84SFMRR117 pKa = 11.84PFADD121 pKa = 2.98IAKK124 pKa = 9.67KK125 pKa = 9.89QLQKK129 pKa = 10.8RR130 pKa = 11.84GVDD133 pKa = 4.0GILARR138 pKa = 11.84KK139 pKa = 8.58YY140 pKa = 10.27PRR142 pKa = 11.84VCRR145 pKa = 11.84GAKK148 pKa = 10.19HH149 pKa = 5.88MGFDD153 pKa = 4.74FNQDD157 pKa = 3.43LQSSEE162 pKa = 4.11LSNSEE167 pKa = 3.64MRR169 pKa = 11.84VKK171 pKa = 10.72QNLGRR176 pKa = 11.84VMFNRR181 pKa = 11.84GLKK184 pKa = 9.07VANLEE189 pKa = 4.18EE190 pKa = 4.39SSSIDD195 pKa = 3.54FEE197 pKa = 4.47
Molecular weight: 22.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2388 |
197 |
1856 |
796.0 |
91.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.276 ± 0.42 | 1.675 ± 0.447 |
7.328 ± 0.217 | 6.826 ± 0.362 |
6.784 ± 0.632 | 5.36 ± 0.249 |
1.424 ± 0.295 | 7.161 ± 0.622 |
9.213 ± 0.843 | 9.757 ± 0.418 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.722 ± 0.586 | 4.983 ± 0.86 |
3.141 ± 0.039 | 2.806 ± 0.31 |
4.899 ± 0.891 | 8.501 ± 0.372 |
2.889 ± 0.298 | 5.318 ± 0.243 |
0.754 ± 0.192 | 3.183 ± 0.474 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
