
Emticicia sp. TH156
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; Emticicia; unclassified Emticicia
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
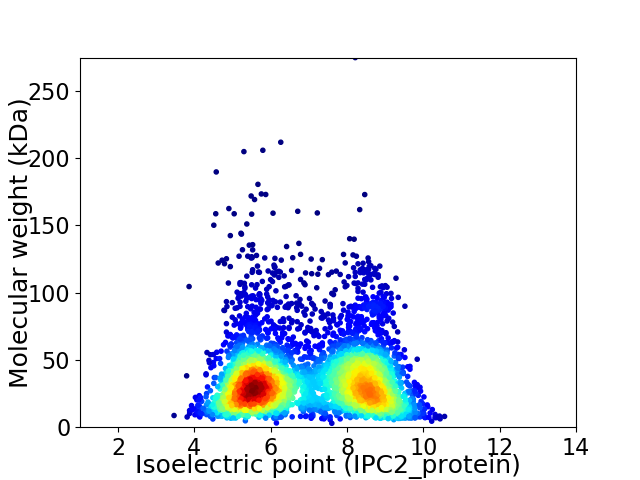
Virtual 2D-PAGE plot for 4502 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N4XDQ6|A0A2N4XDQ6_9BACT RNA polymerase subunit sigma OS=Emticicia sp. TH156 OX=2067454 GN=C0V77_08685 PE=4 SV=1
MM1 pKa = 7.11ITPISFKK8 pKa = 10.81NDD10 pKa = 2.74LSYY13 pKa = 11.26GVSIYY18 pKa = 10.91LSTDD22 pKa = 3.08DD23 pKa = 4.54TNAGQPNYY31 pKa = 9.63IARR34 pKa = 11.84LSFVGHH40 pKa = 6.77IEE42 pKa = 3.88PAATLAISQQSDD54 pKa = 3.46LTSVYY59 pKa = 10.11ICYY62 pKa = 10.39DD63 pKa = 3.54NQNKK67 pKa = 8.48PVKK70 pKa = 10.27RR71 pKa = 11.84MVYY74 pKa = 9.98EE75 pKa = 4.73GGSSQQGFTITQADD89 pKa = 3.69ADD91 pKa = 4.47VIASNYY97 pKa = 10.51SFFAFIAANPNDD109 pKa = 3.94ALSVSFAKK117 pKa = 9.36LTEE120 pKa = 4.69GGNATPDD127 pKa = 3.5QIDD130 pKa = 3.2AFYY133 pKa = 10.62EE134 pKa = 4.42GTANYY139 pKa = 7.83QACTYY144 pKa = 10.41VSYY147 pKa = 11.05ILVLAALSADD157 pKa = 3.76TGTSPADD164 pKa = 3.18TAYY167 pKa = 10.97SLKK170 pKa = 10.99SLVGYY175 pKa = 10.44LGVNWPGLVPDD186 pKa = 4.54FPVRR190 pKa = 11.84SFTCTTLNGNGNLVSLSCDD209 pKa = 2.92VNMEE213 pKa = 4.79DD214 pKa = 5.27IYY216 pKa = 11.19FGSQIAANIFTITPFNSVKK235 pKa = 10.09VVHH238 pKa = 5.95VNISFSYY245 pKa = 9.88TLTPDD250 pKa = 3.56LVTVDD255 pKa = 3.89LTFSLDD261 pKa = 3.58AIRR264 pKa = 11.84IPVAANTDD272 pKa = 3.21ITLSNPAITFSLSPQSGEE290 pKa = 3.9VSFMAQATIPFKK302 pKa = 10.75LWSNPTFYY310 pKa = 11.32ADD312 pKa = 3.17LSMVIGADD320 pKa = 3.21QANIGVVIRR329 pKa = 11.84GDD331 pKa = 3.33NSVLFTPPIIQGVHH345 pKa = 4.4FTSFGVGMGVFFQPIIGYY363 pKa = 8.65SLGVQGSFTIGKK375 pKa = 9.03PEE377 pKa = 4.6DD378 pKa = 3.63NVLVEE383 pKa = 4.91DD384 pKa = 3.61NSFAVVCALIGDD396 pKa = 4.57IPNPLYY402 pKa = 10.36ISFYY406 pKa = 10.49VSQISLNEE414 pKa = 3.85LVTIFTDD421 pKa = 2.95VDD423 pKa = 4.1FNIDD427 pKa = 3.52FPVSVKK433 pKa = 10.52NLSFVWQEE441 pKa = 3.84NPTQPVVLPDD451 pKa = 3.89GSTSKK456 pKa = 11.47YY457 pKa = 9.18EE458 pKa = 4.14FGFSGYY464 pKa = 10.8LDD466 pKa = 3.61FFGLGFYY473 pKa = 11.01GDD475 pKa = 4.88LDD477 pKa = 4.01INPVSGVNGIITMSPYY493 pKa = 10.8EE494 pKa = 4.68LGPLSLKK501 pKa = 11.04GNGQAISIKK510 pKa = 10.32VNEE513 pKa = 4.69AGDD516 pKa = 4.42PISNNVVPEE525 pKa = 4.31TSAEE529 pKa = 3.85EE530 pKa = 3.83LAIEE534 pKa = 4.16NAGTQVIIQAGGPVMEE550 pKa = 4.56VSTFEE555 pKa = 3.84SPYY558 pKa = 9.64FVASIAIAFLDD569 pKa = 3.75CRR571 pKa = 11.84QSVEE575 pKa = 4.41ASVTNNGFSFEE586 pKa = 4.76LDD588 pKa = 3.64FEE590 pKa = 4.54ATLTSSMACVLKK602 pKa = 10.61NYY604 pKa = 10.13QYY606 pKa = 11.19FSGSFTYY613 pKa = 10.44GADD616 pKa = 3.3LMIRR620 pKa = 11.84IPDD623 pKa = 3.67SVDD626 pKa = 3.0VFNLGSIHH634 pKa = 6.24FVSTISSEE642 pKa = 4.17LTIDD646 pKa = 3.75TSADD650 pKa = 3.57VIRR653 pKa = 11.84FTYY656 pKa = 10.42AGSFDD661 pKa = 3.49FAGYY665 pKa = 9.72HH666 pKa = 4.15YY667 pKa = 9.96HH668 pKa = 7.53IPEE671 pKa = 4.71LSLDD675 pKa = 3.59INTQSLQDD683 pKa = 4.01IINTANQWIIDD694 pKa = 3.9NVSTLFKK701 pKa = 10.77NLYY704 pKa = 10.35ADD706 pKa = 3.48ATAWINAITQGVIVLATNTAQYY728 pKa = 10.34VIEE731 pKa = 4.44GFKK734 pKa = 10.67VVYY737 pKa = 9.77GVSAEE742 pKa = 4.11AVGDD746 pKa = 3.86LLKK749 pKa = 10.3GTNYY753 pKa = 10.96ALNDD757 pKa = 3.39VAKK760 pKa = 10.58GMKK763 pKa = 9.58DD764 pKa = 3.29VYY766 pKa = 10.92NATSTLTTQTLVTAYY781 pKa = 10.54NAAEE785 pKa = 4.4DD786 pKa = 3.82SAAAALQYY794 pKa = 11.11AGYY797 pKa = 7.64TADD800 pKa = 4.62EE801 pKa = 4.32IADD804 pKa = 3.74ALVNVYY810 pKa = 10.12GSTMEE815 pKa = 4.07VVGEE819 pKa = 3.82ILLALGYY826 pKa = 10.1SADD829 pKa = 3.66VVAQALTDD837 pKa = 3.59AFNATEE843 pKa = 4.27AEE845 pKa = 4.56VAAVMAEE852 pKa = 4.17LGYY855 pKa = 11.02AIDD858 pKa = 3.77TAGNWVEE865 pKa = 4.41KK866 pKa = 8.29TAKK869 pKa = 10.24DD870 pKa = 3.61AADD873 pKa = 3.59ATKK876 pKa = 10.77NAADD880 pKa = 4.53DD881 pKa = 4.25ALKK884 pKa = 10.56QATDD888 pKa = 3.53STKK891 pKa = 10.95DD892 pKa = 3.37ATDD895 pKa = 3.36SAIKK899 pKa = 8.92EE900 pKa = 4.48TEE902 pKa = 4.06DD903 pKa = 3.5AAKK906 pKa = 10.53DD907 pKa = 3.47AADD910 pKa = 3.93EE911 pKa = 4.33AEE913 pKa = 4.54KK914 pKa = 10.54AAKK917 pKa = 9.99DD918 pKa = 3.4AADD921 pKa = 3.82AAKK924 pKa = 10.49DD925 pKa = 3.62AADD928 pKa = 3.78SALKK932 pKa = 10.56ASEE935 pKa = 4.74DD936 pKa = 3.79AAKK939 pKa = 10.57DD940 pKa = 3.35AADD943 pKa = 3.86AAKK946 pKa = 10.55DD947 pKa = 3.67AADD950 pKa = 4.01EE951 pKa = 4.07AAKK954 pKa = 10.15AAKK957 pKa = 9.96DD958 pKa = 3.34AADD961 pKa = 3.65AAKK964 pKa = 10.53NATDD968 pKa = 3.57SAKK971 pKa = 10.68NATDD975 pKa = 3.53SALKK979 pKa = 10.31SAADD983 pKa = 4.18AIIPP987 pKa = 3.89
MM1 pKa = 7.11ITPISFKK8 pKa = 10.81NDD10 pKa = 2.74LSYY13 pKa = 11.26GVSIYY18 pKa = 10.91LSTDD22 pKa = 3.08DD23 pKa = 4.54TNAGQPNYY31 pKa = 9.63IARR34 pKa = 11.84LSFVGHH40 pKa = 6.77IEE42 pKa = 3.88PAATLAISQQSDD54 pKa = 3.46LTSVYY59 pKa = 10.11ICYY62 pKa = 10.39DD63 pKa = 3.54NQNKK67 pKa = 8.48PVKK70 pKa = 10.27RR71 pKa = 11.84MVYY74 pKa = 9.98EE75 pKa = 4.73GGSSQQGFTITQADD89 pKa = 3.69ADD91 pKa = 4.47VIASNYY97 pKa = 10.51SFFAFIAANPNDD109 pKa = 3.94ALSVSFAKK117 pKa = 9.36LTEE120 pKa = 4.69GGNATPDD127 pKa = 3.5QIDD130 pKa = 3.2AFYY133 pKa = 10.62EE134 pKa = 4.42GTANYY139 pKa = 7.83QACTYY144 pKa = 10.41VSYY147 pKa = 11.05ILVLAALSADD157 pKa = 3.76TGTSPADD164 pKa = 3.18TAYY167 pKa = 10.97SLKK170 pKa = 10.99SLVGYY175 pKa = 10.44LGVNWPGLVPDD186 pKa = 4.54FPVRR190 pKa = 11.84SFTCTTLNGNGNLVSLSCDD209 pKa = 2.92VNMEE213 pKa = 4.79DD214 pKa = 5.27IYY216 pKa = 11.19FGSQIAANIFTITPFNSVKK235 pKa = 10.09VVHH238 pKa = 5.95VNISFSYY245 pKa = 9.88TLTPDD250 pKa = 3.56LVTVDD255 pKa = 3.89LTFSLDD261 pKa = 3.58AIRR264 pKa = 11.84IPVAANTDD272 pKa = 3.21ITLSNPAITFSLSPQSGEE290 pKa = 3.9VSFMAQATIPFKK302 pKa = 10.75LWSNPTFYY310 pKa = 11.32ADD312 pKa = 3.17LSMVIGADD320 pKa = 3.21QANIGVVIRR329 pKa = 11.84GDD331 pKa = 3.33NSVLFTPPIIQGVHH345 pKa = 4.4FTSFGVGMGVFFQPIIGYY363 pKa = 8.65SLGVQGSFTIGKK375 pKa = 9.03PEE377 pKa = 4.6DD378 pKa = 3.63NVLVEE383 pKa = 4.91DD384 pKa = 3.61NSFAVVCALIGDD396 pKa = 4.57IPNPLYY402 pKa = 10.36ISFYY406 pKa = 10.49VSQISLNEE414 pKa = 3.85LVTIFTDD421 pKa = 2.95VDD423 pKa = 4.1FNIDD427 pKa = 3.52FPVSVKK433 pKa = 10.52NLSFVWQEE441 pKa = 3.84NPTQPVVLPDD451 pKa = 3.89GSTSKK456 pKa = 11.47YY457 pKa = 9.18EE458 pKa = 4.14FGFSGYY464 pKa = 10.8LDD466 pKa = 3.61FFGLGFYY473 pKa = 11.01GDD475 pKa = 4.88LDD477 pKa = 4.01INPVSGVNGIITMSPYY493 pKa = 10.8EE494 pKa = 4.68LGPLSLKK501 pKa = 11.04GNGQAISIKK510 pKa = 10.32VNEE513 pKa = 4.69AGDD516 pKa = 4.42PISNNVVPEE525 pKa = 4.31TSAEE529 pKa = 3.85EE530 pKa = 3.83LAIEE534 pKa = 4.16NAGTQVIIQAGGPVMEE550 pKa = 4.56VSTFEE555 pKa = 3.84SPYY558 pKa = 9.64FVASIAIAFLDD569 pKa = 3.75CRR571 pKa = 11.84QSVEE575 pKa = 4.41ASVTNNGFSFEE586 pKa = 4.76LDD588 pKa = 3.64FEE590 pKa = 4.54ATLTSSMACVLKK602 pKa = 10.61NYY604 pKa = 10.13QYY606 pKa = 11.19FSGSFTYY613 pKa = 10.44GADD616 pKa = 3.3LMIRR620 pKa = 11.84IPDD623 pKa = 3.67SVDD626 pKa = 3.0VFNLGSIHH634 pKa = 6.24FVSTISSEE642 pKa = 4.17LTIDD646 pKa = 3.75TSADD650 pKa = 3.57VIRR653 pKa = 11.84FTYY656 pKa = 10.42AGSFDD661 pKa = 3.49FAGYY665 pKa = 9.72HH666 pKa = 4.15YY667 pKa = 9.96HH668 pKa = 7.53IPEE671 pKa = 4.71LSLDD675 pKa = 3.59INTQSLQDD683 pKa = 4.01IINTANQWIIDD694 pKa = 3.9NVSTLFKK701 pKa = 10.77NLYY704 pKa = 10.35ADD706 pKa = 3.48ATAWINAITQGVIVLATNTAQYY728 pKa = 10.34VIEE731 pKa = 4.44GFKK734 pKa = 10.67VVYY737 pKa = 9.77GVSAEE742 pKa = 4.11AVGDD746 pKa = 3.86LLKK749 pKa = 10.3GTNYY753 pKa = 10.96ALNDD757 pKa = 3.39VAKK760 pKa = 10.58GMKK763 pKa = 9.58DD764 pKa = 3.29VYY766 pKa = 10.92NATSTLTTQTLVTAYY781 pKa = 10.54NAAEE785 pKa = 4.4DD786 pKa = 3.82SAAAALQYY794 pKa = 11.11AGYY797 pKa = 7.64TADD800 pKa = 4.62EE801 pKa = 4.32IADD804 pKa = 3.74ALVNVYY810 pKa = 10.12GSTMEE815 pKa = 4.07VVGEE819 pKa = 3.82ILLALGYY826 pKa = 10.1SADD829 pKa = 3.66VVAQALTDD837 pKa = 3.59AFNATEE843 pKa = 4.27AEE845 pKa = 4.56VAAVMAEE852 pKa = 4.17LGYY855 pKa = 11.02AIDD858 pKa = 3.77TAGNWVEE865 pKa = 4.41KK866 pKa = 8.29TAKK869 pKa = 10.24DD870 pKa = 3.61AADD873 pKa = 3.59ATKK876 pKa = 10.77NAADD880 pKa = 4.53DD881 pKa = 4.25ALKK884 pKa = 10.56QATDD888 pKa = 3.53STKK891 pKa = 10.95DD892 pKa = 3.37ATDD895 pKa = 3.36SAIKK899 pKa = 8.92EE900 pKa = 4.48TEE902 pKa = 4.06DD903 pKa = 3.5AAKK906 pKa = 10.53DD907 pKa = 3.47AADD910 pKa = 3.93EE911 pKa = 4.33AEE913 pKa = 4.54KK914 pKa = 10.54AAKK917 pKa = 9.99DD918 pKa = 3.4AADD921 pKa = 3.82AAKK924 pKa = 10.49DD925 pKa = 3.62AADD928 pKa = 3.78SALKK932 pKa = 10.56ASEE935 pKa = 4.74DD936 pKa = 3.79AAKK939 pKa = 10.57DD940 pKa = 3.35AADD943 pKa = 3.86AAKK946 pKa = 10.55DD947 pKa = 3.67AADD950 pKa = 4.01EE951 pKa = 4.07AAKK954 pKa = 10.15AAKK957 pKa = 9.96DD958 pKa = 3.34AADD961 pKa = 3.65AAKK964 pKa = 10.53NATDD968 pKa = 3.57SAKK971 pKa = 10.68NATDD975 pKa = 3.53SALKK979 pKa = 10.31SAADD983 pKa = 4.18AIIPP987 pKa = 3.89
Molecular weight: 104.57 kDa
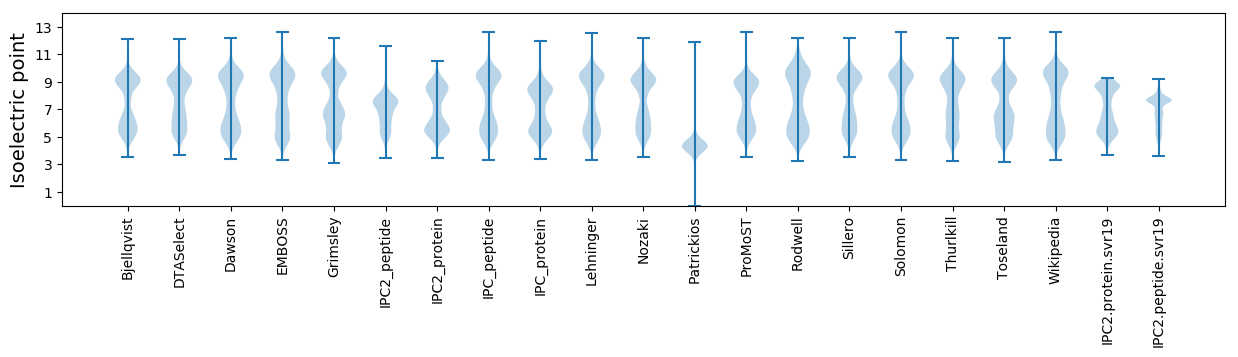
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N4X6Q9|A0A2N4X6Q9_9BACT TIR domain-containing protein OS=Emticicia sp. TH156 OX=2067454 GN=C0V77_22715 PE=4 SV=1
MM1 pKa = 7.92RR2 pKa = 11.84ILRR5 pKa = 11.84FTLFFFIAGLLIAPQVQATHH25 pKa = 6.19LRR27 pKa = 11.84AGEE30 pKa = 3.92ITAKK34 pKa = 10.22RR35 pKa = 11.84ISSTTLTYY43 pKa = 10.55RR44 pKa = 11.84VTLTTYY50 pKa = 9.08TDD52 pKa = 3.46QINGYY57 pKa = 8.6IANDD61 pKa = 3.62AANTSQFSFGVTGVPLFEE79 pKa = 4.15VKK81 pKa = 9.8RR82 pKa = 11.84RR83 pKa = 11.84KK84 pKa = 9.91KK85 pKa = 10.62FLINSS90 pKa = 3.94
MM1 pKa = 7.92RR2 pKa = 11.84ILRR5 pKa = 11.84FTLFFFIAGLLIAPQVQATHH25 pKa = 6.19LRR27 pKa = 11.84AGEE30 pKa = 3.92ITAKK34 pKa = 10.22RR35 pKa = 11.84ISSTTLTYY43 pKa = 10.55RR44 pKa = 11.84VTLTTYY50 pKa = 9.08TDD52 pKa = 3.46QINGYY57 pKa = 8.6IANDD61 pKa = 3.62AANTSQFSFGVTGVPLFEE79 pKa = 4.15VKK81 pKa = 9.8RR82 pKa = 11.84RR83 pKa = 11.84KK84 pKa = 9.91KK85 pKa = 10.62FLINSS90 pKa = 3.94
Molecular weight: 10.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1548216 |
25 |
2417 |
343.9 |
38.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.435 ± 0.037 | 0.775 ± 0.011 |
5.155 ± 0.024 | 6.077 ± 0.041 |
5.089 ± 0.032 | 6.764 ± 0.035 |
1.791 ± 0.017 | 7.226 ± 0.035 |
7.186 ± 0.032 | 9.508 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.338 ± 0.018 | 5.51 ± 0.032 |
3.866 ± 0.021 | 3.726 ± 0.023 |
4.149 ± 0.024 | 6.108 ± 0.025 |
5.655 ± 0.026 | 6.377 ± 0.026 |
1.225 ± 0.015 | 4.041 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
