
Allium cepa amalgavirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; Amalgavirus
Average proteome isoelectric point is 8.28
Get precalculated fractions of proteins
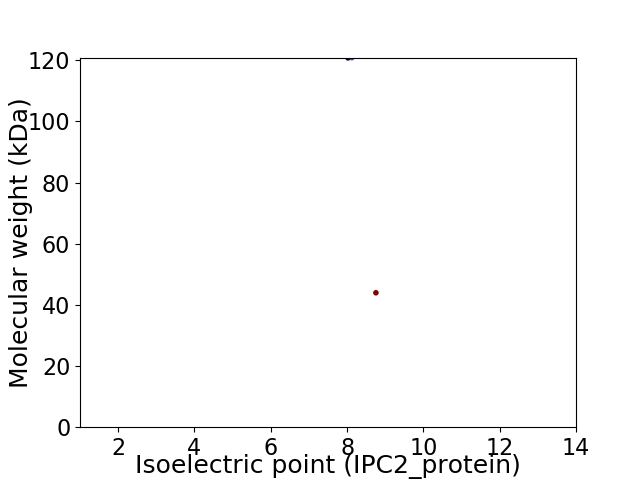
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
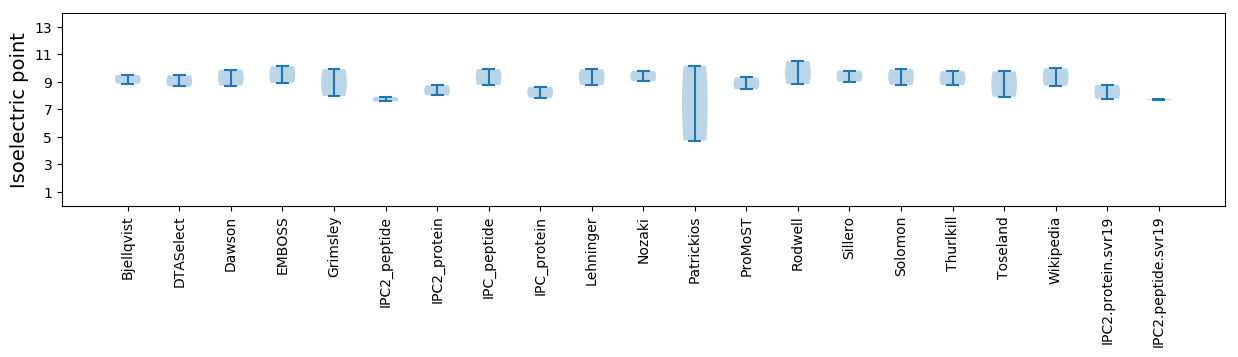
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5MI89|A0A2H5MI89_9VIRU ORF1+2p OS=Allium cepa amalgavirus 1 OX=2058778 GN=ORF1+ORF2 PE=4 SV=1
MM1 pKa = 7.2SAEE4 pKa = 4.36DD5 pKa = 3.62FTPRR9 pKa = 11.84TGLVDD14 pKa = 3.47TEE16 pKa = 4.67LLVDD20 pKa = 4.16PAAEE24 pKa = 4.17LEE26 pKa = 4.19EE27 pKa = 4.3LHH29 pKa = 6.95DD30 pKa = 4.4ALMPLEE36 pKa = 3.97VLNINVKK43 pKa = 10.51AFTRR47 pKa = 11.84ANIFALRR54 pKa = 11.84MTVPQYY60 pKa = 10.2IKK62 pKa = 10.34EE63 pKa = 4.11VRR65 pKa = 11.84VLQNLQDD72 pKa = 3.46VQLIRR77 pKa = 11.84KK78 pKa = 7.73VWTEE82 pKa = 4.16AISHH86 pKa = 6.23KK87 pKa = 10.57CATLPDD93 pKa = 3.72EE94 pKa = 5.3ADD96 pKa = 3.38AGVCLSFARR105 pKa = 11.84WLKK108 pKa = 10.28RR109 pKa = 11.84GVGAKK114 pKa = 9.72VLSDD118 pKa = 3.41AQHH121 pKa = 7.03LIKK124 pKa = 10.47LQKK127 pKa = 9.77RR128 pKa = 11.84AVGSVEE134 pKa = 3.98PKK136 pKa = 10.37VLAFVQLLDD145 pKa = 3.54QQIADD150 pKa = 3.41MHH152 pKa = 6.61AEE154 pKa = 3.97RR155 pKa = 11.84KK156 pKa = 9.6KK157 pKa = 10.57IQAEE161 pKa = 4.31GQAKK165 pKa = 9.25IDD167 pKa = 3.5EE168 pKa = 4.57LRR170 pKa = 11.84RR171 pKa = 11.84EE172 pKa = 3.99IQRR175 pKa = 11.84VEE177 pKa = 3.68RR178 pKa = 11.84EE179 pKa = 3.71YY180 pKa = 11.37DD181 pKa = 3.49EE182 pKa = 4.47KK183 pKa = 11.52AKK185 pKa = 10.86ASSKK189 pKa = 9.92RR190 pKa = 11.84FKK192 pKa = 10.15PARR195 pKa = 11.84KK196 pKa = 9.02YY197 pKa = 10.82VPPTKK202 pKa = 10.62AKK204 pKa = 9.87IDD206 pKa = 3.83EE207 pKa = 4.6EE208 pKa = 4.39CWNAYY213 pKa = 8.88LNKK216 pKa = 9.32VQKK219 pKa = 10.16SGKK222 pKa = 7.22TAPEE226 pKa = 3.73WNAVLQEE233 pKa = 4.23QANTMYY239 pKa = 10.32QQLYY243 pKa = 8.14LTQHH247 pKa = 5.94KK248 pKa = 10.46QDD250 pKa = 3.44FCGLEE255 pKa = 4.21SNQIPLKK262 pKa = 10.33VWADD266 pKa = 3.5SKK268 pKa = 11.37LKK270 pKa = 10.45EE271 pKa = 4.19LADD274 pKa = 3.36NHH276 pKa = 6.15EE277 pKa = 4.96FVASARR283 pKa = 11.84PSEE286 pKa = 4.35SWVSRR291 pKa = 11.84VEE293 pKa = 4.17SHH295 pKa = 6.7LHH297 pKa = 6.29RR298 pKa = 11.84LPLPQRR304 pKa = 11.84VFQANNVPVGPVSQLVMNQSRR325 pKa = 11.84NMALRR330 pKa = 11.84TLLNPQLLTWKK341 pKa = 9.9SQIGMRR347 pKa = 11.84KK348 pKa = 9.42KK349 pKa = 10.56PPLQKK354 pKa = 10.72ANEE357 pKa = 4.19SRR359 pKa = 11.84SSKK362 pKa = 10.23IDD364 pKa = 3.46LLLRR368 pKa = 11.84NPRR371 pKa = 11.84ILVLRR376 pKa = 11.84KK377 pKa = 9.39PEE379 pKa = 4.29RR380 pKa = 11.84GPLGGIPTARR390 pKa = 11.84SKK392 pKa = 11.21FEE394 pKa = 3.76AMVRR398 pKa = 11.84KK399 pKa = 10.0VIGGGEE405 pKa = 4.02MLNWSIDD412 pKa = 3.41SNMYY416 pKa = 10.3RR417 pKa = 11.84GGGNFTDD424 pKa = 4.67ALTLLADD431 pKa = 3.8ARR433 pKa = 11.84YY434 pKa = 8.4DD435 pKa = 3.55APEE438 pKa = 4.26MFLSDD443 pKa = 3.6YY444 pKa = 11.35LSIEE448 pKa = 4.09KK449 pKa = 10.34ARR451 pKa = 11.84SLLCLPSDD459 pKa = 4.32LKK461 pKa = 11.38VPCHH465 pKa = 6.22RR466 pKa = 11.84NCVSVNNFNNEE477 pKa = 3.37ATAGPFFRR485 pKa = 11.84AHH487 pKa = 7.73GIRR490 pKa = 11.84NKK492 pKa = 10.74YY493 pKa = 9.14GMRR496 pKa = 11.84LQLEE500 pKa = 4.23DD501 pKa = 4.92FAWEE505 pKa = 4.55CYY507 pKa = 8.84NAYY510 pKa = 9.97VDD512 pKa = 4.35SGGNPSMLPYY522 pKa = 10.06ISSRR526 pKa = 11.84VGFRR530 pKa = 11.84TKK532 pKa = 10.53LVSTTEE538 pKa = 4.01AFAKK542 pKa = 10.25MKK544 pKa = 10.26DD545 pKa = 3.32NKK547 pKa = 10.54PIGRR551 pKa = 11.84CVMMLDD557 pKa = 4.91AIEE560 pKa = 4.41QMFSTPLYY568 pKa = 10.03NVLSKK573 pKa = 8.07TTAAQRR579 pKa = 11.84FDD581 pKa = 3.23IKK583 pKa = 11.04SGFRR587 pKa = 11.84NTIVRR592 pKa = 11.84ASSDD596 pKa = 2.73WAKK599 pKa = 10.46FWEE602 pKa = 4.42EE603 pKa = 3.56VKK605 pKa = 10.31AAKK608 pKa = 10.42VIVEE612 pKa = 4.28LDD614 pKa = 2.88WKK616 pKa = 10.85KK617 pKa = 10.69FDD619 pKa = 4.23RR620 pKa = 11.84EE621 pKa = 4.11RR622 pKa = 11.84PTEE625 pKa = 4.05DD626 pKa = 3.74LEE628 pKa = 5.5FIIQVIISCFKK639 pKa = 9.55PTDD642 pKa = 3.27EE643 pKa = 5.36RR644 pKa = 11.84EE645 pKa = 3.89RR646 pKa = 11.84LFLRR650 pKa = 11.84GYY652 pKa = 10.81GIMLRR657 pKa = 11.84RR658 pKa = 11.84CLIDD662 pKa = 4.24RR663 pKa = 11.84YY664 pKa = 10.44FITDD668 pKa = 3.36DD669 pKa = 3.69GGVFKK674 pKa = 10.44IDD676 pKa = 3.25GMVPSGSLWTGWVDD690 pKa = 3.2TALNILYY697 pKa = 8.69LQSVMISLGLVSTAVSPKK715 pKa = 10.4CAGDD719 pKa = 4.0DD720 pKa = 3.7NLTLFWKK727 pKa = 10.68DD728 pKa = 3.18FDD730 pKa = 4.59DD731 pKa = 5.82KK732 pKa = 11.31RR733 pKa = 11.84LLSIKK738 pKa = 9.31TRR740 pKa = 11.84LNAWFRR746 pKa = 11.84AGIDD750 pKa = 3.69DD751 pKa = 4.04EE752 pKa = 5.82DD753 pKa = 4.81FFIHH757 pKa = 6.78RR758 pKa = 11.84PPYY761 pKa = 9.97HH762 pKa = 5.2VTRR765 pKa = 11.84EE766 pKa = 4.01QATFPPGTDD775 pKa = 3.16LTKK778 pKa = 10.06GTSRR782 pKa = 11.84KK783 pKa = 9.96LKK785 pKa = 9.73DD786 pKa = 4.7AIWIPIDD793 pKa = 3.61GEE795 pKa = 4.34PIIDD799 pKa = 3.55QAQGLSHH806 pKa = 6.84RR807 pKa = 11.84WQYY810 pKa = 9.82VFRR813 pKa = 11.84GKK815 pKa = 9.77PKK817 pKa = 10.19FLSAYY822 pKa = 7.8WLEE825 pKa = 4.52DD826 pKa = 3.06GRR828 pKa = 11.84PIRR831 pKa = 11.84PTHH834 pKa = 5.8VNAEE838 pKa = 4.07KK839 pKa = 10.92LLFPEE844 pKa = 6.28GIHH847 pKa = 7.16KK848 pKa = 10.51DD849 pKa = 3.04IEE851 pKa = 4.43QYY853 pKa = 10.82EE854 pKa = 4.52SAVLSMIVDD863 pKa = 3.64NPYY866 pKa = 9.65NHH868 pKa = 7.02HH869 pKa = 6.21NVNHH873 pKa = 6.0CMHH876 pKa = 6.99RR877 pKa = 11.84FIICEE882 pKa = 3.71QVKK885 pKa = 9.35RR886 pKa = 11.84QSAAGIDD893 pKa = 3.71PVDD896 pKa = 3.48ILWFSRR902 pKa = 11.84IRR904 pKa = 11.84PGNPDD909 pKa = 3.39VVPYY913 pKa = 10.6PMVASWRR920 pKa = 11.84RR921 pKa = 11.84CEE923 pKa = 4.44GYY925 pKa = 10.74VDD927 pKa = 5.38LEE929 pKa = 3.95QLPFISDD936 pKa = 3.53YY937 pKa = 11.71VKK939 pKa = 10.7DD940 pKa = 3.68LKK942 pKa = 11.18EE943 pKa = 3.96FVAGVTSLYY952 pKa = 10.99ARR954 pKa = 11.84DD955 pKa = 4.0SIGGLDD961 pKa = 2.97AWRR964 pKa = 11.84FTEE967 pKa = 4.72IIRR970 pKa = 11.84GEE972 pKa = 4.15TDD974 pKa = 2.8LGTGQFGNDD983 pKa = 2.52IDD985 pKa = 4.86VWIHH989 pKa = 5.62WLHH992 pKa = 5.91HH993 pKa = 6.38HH994 pKa = 7.24PLTKK998 pKa = 9.92YY999 pKa = 9.33LKK1001 pKa = 8.78PIKK1004 pKa = 9.86RR1005 pKa = 11.84HH1006 pKa = 4.96RR1007 pKa = 11.84QPPLSVMPDD1016 pKa = 3.22KK1017 pKa = 10.46EE1018 pKa = 3.91LHH1020 pKa = 5.53GRR1022 pKa = 11.84IEE1024 pKa = 4.04EE1025 pKa = 4.14ALQVYY1030 pKa = 10.12RR1031 pKa = 11.84DD1032 pKa = 3.53IRR1034 pKa = 11.84GTTTNASAEE1043 pKa = 4.11AFSLFLSNVIKK1054 pKa = 10.55HH1055 pKa = 5.57QGG1057 pKa = 2.91
MM1 pKa = 7.2SAEE4 pKa = 4.36DD5 pKa = 3.62FTPRR9 pKa = 11.84TGLVDD14 pKa = 3.47TEE16 pKa = 4.67LLVDD20 pKa = 4.16PAAEE24 pKa = 4.17LEE26 pKa = 4.19EE27 pKa = 4.3LHH29 pKa = 6.95DD30 pKa = 4.4ALMPLEE36 pKa = 3.97VLNINVKK43 pKa = 10.51AFTRR47 pKa = 11.84ANIFALRR54 pKa = 11.84MTVPQYY60 pKa = 10.2IKK62 pKa = 10.34EE63 pKa = 4.11VRR65 pKa = 11.84VLQNLQDD72 pKa = 3.46VQLIRR77 pKa = 11.84KK78 pKa = 7.73VWTEE82 pKa = 4.16AISHH86 pKa = 6.23KK87 pKa = 10.57CATLPDD93 pKa = 3.72EE94 pKa = 5.3ADD96 pKa = 3.38AGVCLSFARR105 pKa = 11.84WLKK108 pKa = 10.28RR109 pKa = 11.84GVGAKK114 pKa = 9.72VLSDD118 pKa = 3.41AQHH121 pKa = 7.03LIKK124 pKa = 10.47LQKK127 pKa = 9.77RR128 pKa = 11.84AVGSVEE134 pKa = 3.98PKK136 pKa = 10.37VLAFVQLLDD145 pKa = 3.54QQIADD150 pKa = 3.41MHH152 pKa = 6.61AEE154 pKa = 3.97RR155 pKa = 11.84KK156 pKa = 9.6KK157 pKa = 10.57IQAEE161 pKa = 4.31GQAKK165 pKa = 9.25IDD167 pKa = 3.5EE168 pKa = 4.57LRR170 pKa = 11.84RR171 pKa = 11.84EE172 pKa = 3.99IQRR175 pKa = 11.84VEE177 pKa = 3.68RR178 pKa = 11.84EE179 pKa = 3.71YY180 pKa = 11.37DD181 pKa = 3.49EE182 pKa = 4.47KK183 pKa = 11.52AKK185 pKa = 10.86ASSKK189 pKa = 9.92RR190 pKa = 11.84FKK192 pKa = 10.15PARR195 pKa = 11.84KK196 pKa = 9.02YY197 pKa = 10.82VPPTKK202 pKa = 10.62AKK204 pKa = 9.87IDD206 pKa = 3.83EE207 pKa = 4.6EE208 pKa = 4.39CWNAYY213 pKa = 8.88LNKK216 pKa = 9.32VQKK219 pKa = 10.16SGKK222 pKa = 7.22TAPEE226 pKa = 3.73WNAVLQEE233 pKa = 4.23QANTMYY239 pKa = 10.32QQLYY243 pKa = 8.14LTQHH247 pKa = 5.94KK248 pKa = 10.46QDD250 pKa = 3.44FCGLEE255 pKa = 4.21SNQIPLKK262 pKa = 10.33VWADD266 pKa = 3.5SKK268 pKa = 11.37LKK270 pKa = 10.45EE271 pKa = 4.19LADD274 pKa = 3.36NHH276 pKa = 6.15EE277 pKa = 4.96FVASARR283 pKa = 11.84PSEE286 pKa = 4.35SWVSRR291 pKa = 11.84VEE293 pKa = 4.17SHH295 pKa = 6.7LHH297 pKa = 6.29RR298 pKa = 11.84LPLPQRR304 pKa = 11.84VFQANNVPVGPVSQLVMNQSRR325 pKa = 11.84NMALRR330 pKa = 11.84TLLNPQLLTWKK341 pKa = 9.9SQIGMRR347 pKa = 11.84KK348 pKa = 9.42KK349 pKa = 10.56PPLQKK354 pKa = 10.72ANEE357 pKa = 4.19SRR359 pKa = 11.84SSKK362 pKa = 10.23IDD364 pKa = 3.46LLLRR368 pKa = 11.84NPRR371 pKa = 11.84ILVLRR376 pKa = 11.84KK377 pKa = 9.39PEE379 pKa = 4.29RR380 pKa = 11.84GPLGGIPTARR390 pKa = 11.84SKK392 pKa = 11.21FEE394 pKa = 3.76AMVRR398 pKa = 11.84KK399 pKa = 10.0VIGGGEE405 pKa = 4.02MLNWSIDD412 pKa = 3.41SNMYY416 pKa = 10.3RR417 pKa = 11.84GGGNFTDD424 pKa = 4.67ALTLLADD431 pKa = 3.8ARR433 pKa = 11.84YY434 pKa = 8.4DD435 pKa = 3.55APEE438 pKa = 4.26MFLSDD443 pKa = 3.6YY444 pKa = 11.35LSIEE448 pKa = 4.09KK449 pKa = 10.34ARR451 pKa = 11.84SLLCLPSDD459 pKa = 4.32LKK461 pKa = 11.38VPCHH465 pKa = 6.22RR466 pKa = 11.84NCVSVNNFNNEE477 pKa = 3.37ATAGPFFRR485 pKa = 11.84AHH487 pKa = 7.73GIRR490 pKa = 11.84NKK492 pKa = 10.74YY493 pKa = 9.14GMRR496 pKa = 11.84LQLEE500 pKa = 4.23DD501 pKa = 4.92FAWEE505 pKa = 4.55CYY507 pKa = 8.84NAYY510 pKa = 9.97VDD512 pKa = 4.35SGGNPSMLPYY522 pKa = 10.06ISSRR526 pKa = 11.84VGFRR530 pKa = 11.84TKK532 pKa = 10.53LVSTTEE538 pKa = 4.01AFAKK542 pKa = 10.25MKK544 pKa = 10.26DD545 pKa = 3.32NKK547 pKa = 10.54PIGRR551 pKa = 11.84CVMMLDD557 pKa = 4.91AIEE560 pKa = 4.41QMFSTPLYY568 pKa = 10.03NVLSKK573 pKa = 8.07TTAAQRR579 pKa = 11.84FDD581 pKa = 3.23IKK583 pKa = 11.04SGFRR587 pKa = 11.84NTIVRR592 pKa = 11.84ASSDD596 pKa = 2.73WAKK599 pKa = 10.46FWEE602 pKa = 4.42EE603 pKa = 3.56VKK605 pKa = 10.31AAKK608 pKa = 10.42VIVEE612 pKa = 4.28LDD614 pKa = 2.88WKK616 pKa = 10.85KK617 pKa = 10.69FDD619 pKa = 4.23RR620 pKa = 11.84EE621 pKa = 4.11RR622 pKa = 11.84PTEE625 pKa = 4.05DD626 pKa = 3.74LEE628 pKa = 5.5FIIQVIISCFKK639 pKa = 9.55PTDD642 pKa = 3.27EE643 pKa = 5.36RR644 pKa = 11.84EE645 pKa = 3.89RR646 pKa = 11.84LFLRR650 pKa = 11.84GYY652 pKa = 10.81GIMLRR657 pKa = 11.84RR658 pKa = 11.84CLIDD662 pKa = 4.24RR663 pKa = 11.84YY664 pKa = 10.44FITDD668 pKa = 3.36DD669 pKa = 3.69GGVFKK674 pKa = 10.44IDD676 pKa = 3.25GMVPSGSLWTGWVDD690 pKa = 3.2TALNILYY697 pKa = 8.69LQSVMISLGLVSTAVSPKK715 pKa = 10.4CAGDD719 pKa = 4.0DD720 pKa = 3.7NLTLFWKK727 pKa = 10.68DD728 pKa = 3.18FDD730 pKa = 4.59DD731 pKa = 5.82KK732 pKa = 11.31RR733 pKa = 11.84LLSIKK738 pKa = 9.31TRR740 pKa = 11.84LNAWFRR746 pKa = 11.84AGIDD750 pKa = 3.69DD751 pKa = 4.04EE752 pKa = 5.82DD753 pKa = 4.81FFIHH757 pKa = 6.78RR758 pKa = 11.84PPYY761 pKa = 9.97HH762 pKa = 5.2VTRR765 pKa = 11.84EE766 pKa = 4.01QATFPPGTDD775 pKa = 3.16LTKK778 pKa = 10.06GTSRR782 pKa = 11.84KK783 pKa = 9.96LKK785 pKa = 9.73DD786 pKa = 4.7AIWIPIDD793 pKa = 3.61GEE795 pKa = 4.34PIIDD799 pKa = 3.55QAQGLSHH806 pKa = 6.84RR807 pKa = 11.84WQYY810 pKa = 9.82VFRR813 pKa = 11.84GKK815 pKa = 9.77PKK817 pKa = 10.19FLSAYY822 pKa = 7.8WLEE825 pKa = 4.52DD826 pKa = 3.06GRR828 pKa = 11.84PIRR831 pKa = 11.84PTHH834 pKa = 5.8VNAEE838 pKa = 4.07KK839 pKa = 10.92LLFPEE844 pKa = 6.28GIHH847 pKa = 7.16KK848 pKa = 10.51DD849 pKa = 3.04IEE851 pKa = 4.43QYY853 pKa = 10.82EE854 pKa = 4.52SAVLSMIVDD863 pKa = 3.64NPYY866 pKa = 9.65NHH868 pKa = 7.02HH869 pKa = 6.21NVNHH873 pKa = 6.0CMHH876 pKa = 6.99RR877 pKa = 11.84FIICEE882 pKa = 3.71QVKK885 pKa = 9.35RR886 pKa = 11.84QSAAGIDD893 pKa = 3.71PVDD896 pKa = 3.48ILWFSRR902 pKa = 11.84IRR904 pKa = 11.84PGNPDD909 pKa = 3.39VVPYY913 pKa = 10.6PMVASWRR920 pKa = 11.84RR921 pKa = 11.84CEE923 pKa = 4.44GYY925 pKa = 10.74VDD927 pKa = 5.38LEE929 pKa = 3.95QLPFISDD936 pKa = 3.53YY937 pKa = 11.71VKK939 pKa = 10.7DD940 pKa = 3.68LKK942 pKa = 11.18EE943 pKa = 3.96FVAGVTSLYY952 pKa = 10.99ARR954 pKa = 11.84DD955 pKa = 4.0SIGGLDD961 pKa = 2.97AWRR964 pKa = 11.84FTEE967 pKa = 4.72IIRR970 pKa = 11.84GEE972 pKa = 4.15TDD974 pKa = 2.8LGTGQFGNDD983 pKa = 2.52IDD985 pKa = 4.86VWIHH989 pKa = 5.62WLHH992 pKa = 5.91HH993 pKa = 6.38HH994 pKa = 7.24PLTKK998 pKa = 9.92YY999 pKa = 9.33LKK1001 pKa = 8.78PIKK1004 pKa = 9.86RR1005 pKa = 11.84HH1006 pKa = 4.96RR1007 pKa = 11.84QPPLSVMPDD1016 pKa = 3.22KK1017 pKa = 10.46EE1018 pKa = 3.91LHH1020 pKa = 5.53GRR1022 pKa = 11.84IEE1024 pKa = 4.04EE1025 pKa = 4.14ALQVYY1030 pKa = 10.12RR1031 pKa = 11.84DD1032 pKa = 3.53IRR1034 pKa = 11.84GTTTNASAEE1043 pKa = 4.11AFSLFLSNVIKK1054 pKa = 10.55HH1055 pKa = 5.57QGG1057 pKa = 2.91
Molecular weight: 120.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5MI89|A0A2H5MI89_9VIRU ORF1+2p OS=Allium cepa amalgavirus 1 OX=2058778 GN=ORF1+ORF2 PE=4 SV=1
MM1 pKa = 7.2SAEE4 pKa = 4.36DD5 pKa = 3.62FTPRR9 pKa = 11.84TGLVDD14 pKa = 3.47TEE16 pKa = 4.67LLVDD20 pKa = 4.16PAAEE24 pKa = 4.17LEE26 pKa = 4.19EE27 pKa = 4.3LHH29 pKa = 6.95DD30 pKa = 4.4ALMPLEE36 pKa = 3.97VLNINVKK43 pKa = 10.51AFTRR47 pKa = 11.84ANIFALRR54 pKa = 11.84MTVPQYY60 pKa = 10.2IKK62 pKa = 10.34EE63 pKa = 4.11VRR65 pKa = 11.84VLQNLQDD72 pKa = 3.46VQLIRR77 pKa = 11.84KK78 pKa = 7.73VWTEE82 pKa = 4.16AISHH86 pKa = 6.23KK87 pKa = 10.57CATLPDD93 pKa = 3.72EE94 pKa = 5.3ADD96 pKa = 3.38AGVCLSFARR105 pKa = 11.84WLKK108 pKa = 10.28RR109 pKa = 11.84GVGAKK114 pKa = 9.72VLSDD118 pKa = 3.41AQHH121 pKa = 7.03LIKK124 pKa = 10.47LQKK127 pKa = 9.77RR128 pKa = 11.84AVGSVEE134 pKa = 3.98PKK136 pKa = 10.37VLAFVQLLDD145 pKa = 3.54QQIADD150 pKa = 3.41MHH152 pKa = 6.61AEE154 pKa = 3.97RR155 pKa = 11.84KK156 pKa = 9.6KK157 pKa = 10.57IQAEE161 pKa = 4.31GQAKK165 pKa = 9.25IDD167 pKa = 3.5EE168 pKa = 4.57LRR170 pKa = 11.84RR171 pKa = 11.84EE172 pKa = 3.99IQRR175 pKa = 11.84VEE177 pKa = 3.68RR178 pKa = 11.84EE179 pKa = 3.71YY180 pKa = 11.37DD181 pKa = 3.49EE182 pKa = 4.47KK183 pKa = 11.52AKK185 pKa = 10.86ASSKK189 pKa = 9.92RR190 pKa = 11.84FKK192 pKa = 10.15PARR195 pKa = 11.84KK196 pKa = 9.02YY197 pKa = 10.82VPPTKK202 pKa = 10.62AKK204 pKa = 9.87IDD206 pKa = 3.83EE207 pKa = 4.6EE208 pKa = 4.39CWNAYY213 pKa = 8.88LNKK216 pKa = 9.32VQKK219 pKa = 10.16SGKK222 pKa = 7.22TAPEE226 pKa = 3.73WNAVLQEE233 pKa = 4.23QANTMYY239 pKa = 10.32QQLYY243 pKa = 8.14LTQHH247 pKa = 5.94KK248 pKa = 10.46QDD250 pKa = 3.44FCGLEE255 pKa = 4.21SNQIPLKK262 pKa = 10.33VWADD266 pKa = 3.5SKK268 pKa = 11.37LKK270 pKa = 10.45EE271 pKa = 4.19LADD274 pKa = 3.27NHH276 pKa = 5.11EE277 pKa = 4.24FRR279 pKa = 11.84RR280 pKa = 11.84VRR282 pKa = 11.84EE283 pKa = 3.95TQRR286 pKa = 11.84ILGLASGVPSSPSASASKK304 pKa = 10.59SLSGKK309 pKa = 8.42QRR311 pKa = 11.84SGGPSQSAGDD321 pKa = 3.94EE322 pKa = 4.08PVEE325 pKa = 4.05EE326 pKa = 4.82HH327 pKa = 6.76GSSNSAKK334 pKa = 9.86PAAPDD339 pKa = 3.38MEE341 pKa = 4.2ITDD344 pKa = 3.81RR345 pKa = 11.84DD346 pKa = 3.76EE347 pKa = 4.72EE348 pKa = 4.41KK349 pKa = 10.35TSSSKK354 pKa = 10.79GKK356 pKa = 10.2RR357 pKa = 11.84KK358 pKa = 9.44QKK360 pKa = 10.84LKK362 pKa = 11.05DD363 pKa = 3.04RR364 pKa = 11.84LAAKK368 pKa = 9.99KK369 pKa = 10.78SKK371 pKa = 10.46DD372 pKa = 3.31LSAKK376 pKa = 8.59KK377 pKa = 8.27TRR379 pKa = 11.84KK380 pKa = 7.94GTIRR384 pKa = 11.84RR385 pKa = 11.84DD386 pKa = 3.39PHH388 pKa = 6.13RR389 pKa = 11.84KK390 pKa = 8.93KK391 pKa = 10.85
MM1 pKa = 7.2SAEE4 pKa = 4.36DD5 pKa = 3.62FTPRR9 pKa = 11.84TGLVDD14 pKa = 3.47TEE16 pKa = 4.67LLVDD20 pKa = 4.16PAAEE24 pKa = 4.17LEE26 pKa = 4.19EE27 pKa = 4.3LHH29 pKa = 6.95DD30 pKa = 4.4ALMPLEE36 pKa = 3.97VLNINVKK43 pKa = 10.51AFTRR47 pKa = 11.84ANIFALRR54 pKa = 11.84MTVPQYY60 pKa = 10.2IKK62 pKa = 10.34EE63 pKa = 4.11VRR65 pKa = 11.84VLQNLQDD72 pKa = 3.46VQLIRR77 pKa = 11.84KK78 pKa = 7.73VWTEE82 pKa = 4.16AISHH86 pKa = 6.23KK87 pKa = 10.57CATLPDD93 pKa = 3.72EE94 pKa = 5.3ADD96 pKa = 3.38AGVCLSFARR105 pKa = 11.84WLKK108 pKa = 10.28RR109 pKa = 11.84GVGAKK114 pKa = 9.72VLSDD118 pKa = 3.41AQHH121 pKa = 7.03LIKK124 pKa = 10.47LQKK127 pKa = 9.77RR128 pKa = 11.84AVGSVEE134 pKa = 3.98PKK136 pKa = 10.37VLAFVQLLDD145 pKa = 3.54QQIADD150 pKa = 3.41MHH152 pKa = 6.61AEE154 pKa = 3.97RR155 pKa = 11.84KK156 pKa = 9.6KK157 pKa = 10.57IQAEE161 pKa = 4.31GQAKK165 pKa = 9.25IDD167 pKa = 3.5EE168 pKa = 4.57LRR170 pKa = 11.84RR171 pKa = 11.84EE172 pKa = 3.99IQRR175 pKa = 11.84VEE177 pKa = 3.68RR178 pKa = 11.84EE179 pKa = 3.71YY180 pKa = 11.37DD181 pKa = 3.49EE182 pKa = 4.47KK183 pKa = 11.52AKK185 pKa = 10.86ASSKK189 pKa = 9.92RR190 pKa = 11.84FKK192 pKa = 10.15PARR195 pKa = 11.84KK196 pKa = 9.02YY197 pKa = 10.82VPPTKK202 pKa = 10.62AKK204 pKa = 9.87IDD206 pKa = 3.83EE207 pKa = 4.6EE208 pKa = 4.39CWNAYY213 pKa = 8.88LNKK216 pKa = 9.32VQKK219 pKa = 10.16SGKK222 pKa = 7.22TAPEE226 pKa = 3.73WNAVLQEE233 pKa = 4.23QANTMYY239 pKa = 10.32QQLYY243 pKa = 8.14LTQHH247 pKa = 5.94KK248 pKa = 10.46QDD250 pKa = 3.44FCGLEE255 pKa = 4.21SNQIPLKK262 pKa = 10.33VWADD266 pKa = 3.5SKK268 pKa = 11.37LKK270 pKa = 10.45EE271 pKa = 4.19LADD274 pKa = 3.27NHH276 pKa = 5.11EE277 pKa = 4.24FRR279 pKa = 11.84RR280 pKa = 11.84VRR282 pKa = 11.84EE283 pKa = 3.95TQRR286 pKa = 11.84ILGLASGVPSSPSASASKK304 pKa = 10.59SLSGKK309 pKa = 8.42QRR311 pKa = 11.84SGGPSQSAGDD321 pKa = 3.94EE322 pKa = 4.08PVEE325 pKa = 4.05EE326 pKa = 4.82HH327 pKa = 6.76GSSNSAKK334 pKa = 9.86PAAPDD339 pKa = 3.38MEE341 pKa = 4.2ITDD344 pKa = 3.81RR345 pKa = 11.84DD346 pKa = 3.76EE347 pKa = 4.72EE348 pKa = 4.41KK349 pKa = 10.35TSSSKK354 pKa = 10.79GKK356 pKa = 10.2RR357 pKa = 11.84KK358 pKa = 9.44QKK360 pKa = 10.84LKK362 pKa = 11.05DD363 pKa = 3.04RR364 pKa = 11.84LAAKK368 pKa = 9.99KK369 pKa = 10.78SKK371 pKa = 10.46DD372 pKa = 3.31LSAKK376 pKa = 8.59KK377 pKa = 8.27TRR379 pKa = 11.84KK380 pKa = 7.94GTIRR384 pKa = 11.84RR385 pKa = 11.84DD386 pKa = 3.39PHH388 pKa = 6.13RR389 pKa = 11.84KK390 pKa = 8.93KK391 pKa = 10.85
Molecular weight: 43.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1448 |
391 |
1057 |
724.0 |
82.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.08 ± 1.134 | 1.312 ± 0.152 |
6.146 ± 0.139 | 6.492 ± 0.758 |
3.66 ± 0.851 | 5.11 ± 0.402 |
2.348 ± 0.159 | 5.387 ± 0.817 |
7.735 ± 1.72 | 9.599 ± 0.207 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.141 ± 0.32 | 3.729 ± 0.483 |
5.18 ± 0.304 | 4.696 ± 0.76 |
6.975 ± 0.037 | 6.285 ± 0.597 |
4.282 ± 0.1 | 6.492 ± 0.186 |
2.003 ± 0.382 | 2.348 ± 0.429 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
