
Ruminococcaceae bacterium YAD3003
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; unclassified Oscillospiraceae
Average proteome isoelectric point is 5.78
Get precalculated fractions of proteins
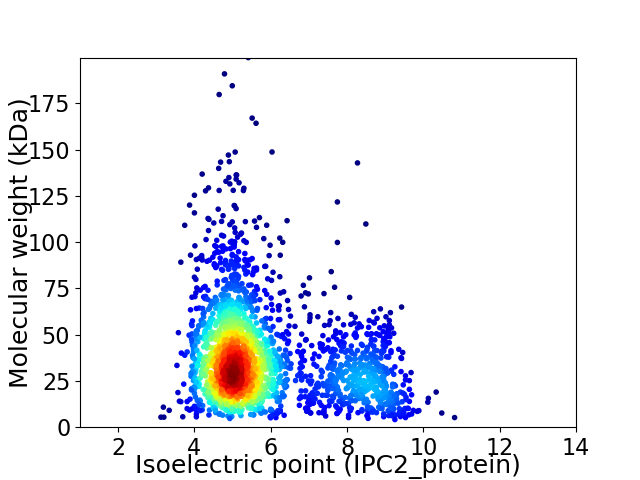
Virtual 2D-PAGE plot for 2342 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3CFU3|A0A1H3CFU3_9FIRM Formate--tetrahydrofolate ligase OS=Ruminococcaceae bacterium YAD3003 OX=1520816 GN=fhs PE=3 SV=1
MM1 pKa = 7.4KK2 pKa = 9.79PVKK5 pKa = 9.74TEE7 pKa = 3.9YY8 pKa = 10.34KK9 pKa = 10.13ALAAITAGTIAVLSIAACSIASSNTPDD36 pKa = 3.07TTVPSEE42 pKa = 4.13SEE44 pKa = 4.4TIVEE48 pKa = 4.55TTFSDD53 pKa = 3.34NDD55 pKa = 3.38LNAGYY60 pKa = 10.32DD61 pKa = 3.36EE62 pKa = 5.48GEE64 pKa = 4.05AQIITLNGTSASSDD78 pKa = 3.25SSSVKK83 pKa = 9.98TEE85 pKa = 3.87GSVVTITGKK94 pKa = 8.55GTYY97 pKa = 10.12VITGTLNDD105 pKa = 4.4GYY107 pKa = 10.5IVVDD111 pKa = 4.61AGDD114 pKa = 3.74SDD116 pKa = 5.4DD117 pKa = 4.8IRR119 pKa = 11.84IVLDD123 pKa = 3.45NADD126 pKa = 3.88ITSSDD131 pKa = 3.5YY132 pKa = 11.55AAIYY136 pKa = 9.55CLNADD141 pKa = 3.42NVYY144 pKa = 8.67ITLADD149 pKa = 3.91GSSNSLSSTGEE160 pKa = 3.72FDD162 pKa = 4.67SKK164 pKa = 11.36DD165 pKa = 3.72SNSVDD170 pKa = 3.01GAIFAKK176 pKa = 10.02TDD178 pKa = 2.99ITINGSGTLKK188 pKa = 10.58INSTDD193 pKa = 3.12HH194 pKa = 6.85GIVGKK199 pKa = 10.35DD200 pKa = 3.15DD201 pKa = 3.33VTITGGGISIKK212 pKa = 10.42SSSDD216 pKa = 3.35GIQANDD222 pKa = 3.4SVSVKK227 pKa = 10.48DD228 pKa = 3.43AVINIVCGKK237 pKa = 10.5DD238 pKa = 3.75GIQADD243 pKa = 3.92NDD245 pKa = 3.63DD246 pKa = 5.27DD247 pKa = 3.88KK248 pKa = 11.65TKK250 pKa = 10.61GYY252 pKa = 11.04VYY254 pKa = 9.97IASGSITISAGDD266 pKa = 3.87DD267 pKa = 4.09GITASSSLQIDD278 pKa = 4.49DD279 pKa = 3.73GTIDD283 pKa = 2.97ITGSYY288 pKa = 9.77EE289 pKa = 4.0GLEE292 pKa = 4.04GRR294 pKa = 11.84YY295 pKa = 7.28VTVNGGTISIVSSDD309 pKa = 3.68DD310 pKa = 3.9GINAVSSTSSGEE322 pKa = 3.75MFQADD327 pKa = 4.48DD328 pKa = 4.75SKK330 pKa = 11.75LCINGGEE337 pKa = 4.36IYY339 pKa = 11.12VNTQGDD345 pKa = 4.42GVDD348 pKa = 3.57SNGTFEE354 pKa = 4.18MTGGILTVMGPTSGANGSLDD374 pKa = 3.73VNGSAVISGGTVVMAGASGMATNFTDD400 pKa = 3.6ATQGTALLTVGNQSEE415 pKa = 4.43GSEE418 pKa = 3.94ITVTDD423 pKa = 3.21STGNVILSATSDD435 pKa = 3.37CSYY438 pKa = 9.97QTVLVSSPDD447 pKa = 3.43MEE449 pKa = 5.76KK450 pKa = 10.95DD451 pKa = 2.94GTYY454 pKa = 9.98TVTAGSYY461 pKa = 10.62SEE463 pKa = 4.73TITLSGYY470 pKa = 10.79LYY472 pKa = 10.72GAGGGFGGGVPGGDD486 pKa = 3.87PGQMPAGNSGDD497 pKa = 3.92FPSGGPGGMHH507 pKa = 7.1PPFF510 pKa = 5.38
MM1 pKa = 7.4KK2 pKa = 9.79PVKK5 pKa = 9.74TEE7 pKa = 3.9YY8 pKa = 10.34KK9 pKa = 10.13ALAAITAGTIAVLSIAACSIASSNTPDD36 pKa = 3.07TTVPSEE42 pKa = 4.13SEE44 pKa = 4.4TIVEE48 pKa = 4.55TTFSDD53 pKa = 3.34NDD55 pKa = 3.38LNAGYY60 pKa = 10.32DD61 pKa = 3.36EE62 pKa = 5.48GEE64 pKa = 4.05AQIITLNGTSASSDD78 pKa = 3.25SSSVKK83 pKa = 9.98TEE85 pKa = 3.87GSVVTITGKK94 pKa = 8.55GTYY97 pKa = 10.12VITGTLNDD105 pKa = 4.4GYY107 pKa = 10.5IVVDD111 pKa = 4.61AGDD114 pKa = 3.74SDD116 pKa = 5.4DD117 pKa = 4.8IRR119 pKa = 11.84IVLDD123 pKa = 3.45NADD126 pKa = 3.88ITSSDD131 pKa = 3.5YY132 pKa = 11.55AAIYY136 pKa = 9.55CLNADD141 pKa = 3.42NVYY144 pKa = 8.67ITLADD149 pKa = 3.91GSSNSLSSTGEE160 pKa = 3.72FDD162 pKa = 4.67SKK164 pKa = 11.36DD165 pKa = 3.72SNSVDD170 pKa = 3.01GAIFAKK176 pKa = 10.02TDD178 pKa = 2.99ITINGSGTLKK188 pKa = 10.58INSTDD193 pKa = 3.12HH194 pKa = 6.85GIVGKK199 pKa = 10.35DD200 pKa = 3.15DD201 pKa = 3.33VTITGGGISIKK212 pKa = 10.42SSSDD216 pKa = 3.35GIQANDD222 pKa = 3.4SVSVKK227 pKa = 10.48DD228 pKa = 3.43AVINIVCGKK237 pKa = 10.5DD238 pKa = 3.75GIQADD243 pKa = 3.92NDD245 pKa = 3.63DD246 pKa = 5.27DD247 pKa = 3.88KK248 pKa = 11.65TKK250 pKa = 10.61GYY252 pKa = 11.04VYY254 pKa = 9.97IASGSITISAGDD266 pKa = 3.87DD267 pKa = 4.09GITASSSLQIDD278 pKa = 4.49DD279 pKa = 3.73GTIDD283 pKa = 2.97ITGSYY288 pKa = 9.77EE289 pKa = 4.0GLEE292 pKa = 4.04GRR294 pKa = 11.84YY295 pKa = 7.28VTVNGGTISIVSSDD309 pKa = 3.68DD310 pKa = 3.9GINAVSSTSSGEE322 pKa = 3.75MFQADD327 pKa = 4.48DD328 pKa = 4.75SKK330 pKa = 11.75LCINGGEE337 pKa = 4.36IYY339 pKa = 11.12VNTQGDD345 pKa = 4.42GVDD348 pKa = 3.57SNGTFEE354 pKa = 4.18MTGGILTVMGPTSGANGSLDD374 pKa = 3.73VNGSAVISGGTVVMAGASGMATNFTDD400 pKa = 3.6ATQGTALLTVGNQSEE415 pKa = 4.43GSEE418 pKa = 3.94ITVTDD423 pKa = 3.21STGNVILSATSDD435 pKa = 3.37CSYY438 pKa = 9.97QTVLVSSPDD447 pKa = 3.43MEE449 pKa = 5.76KK450 pKa = 10.95DD451 pKa = 2.94GTYY454 pKa = 9.98TVTAGSYY461 pKa = 10.62SEE463 pKa = 4.73TITLSGYY470 pKa = 10.79LYY472 pKa = 10.72GAGGGFGGGVPGGDD486 pKa = 3.87PGQMPAGNSGDD497 pKa = 3.92FPSGGPGGMHH507 pKa = 7.1PPFF510 pKa = 5.38
Molecular weight: 51.08 kDa
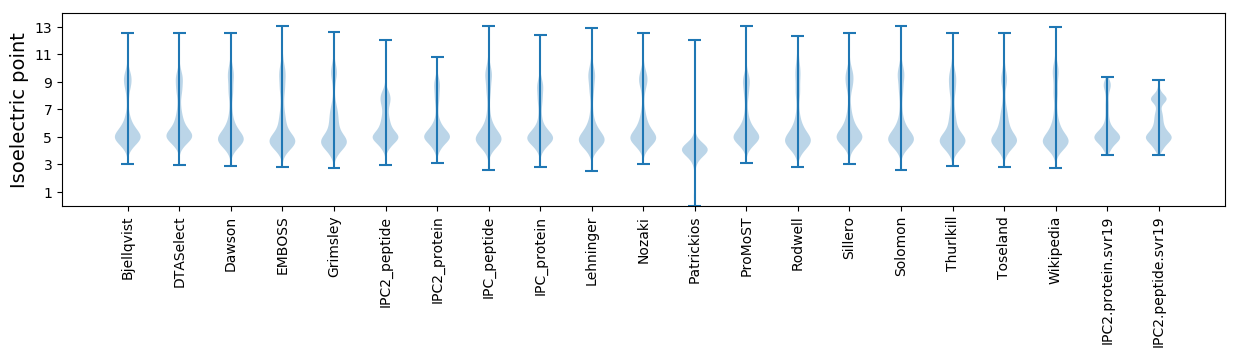
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H3CDP6|A0A1H3CDP6_9FIRM 50S ribosomal protein L16 OS=Ruminococcaceae bacterium YAD3003 OX=1520816 GN=rplP PE=3 SV=1
MM1 pKa = 7.7AKK3 pKa = 8.05MTFQPKK9 pKa = 9.04KK10 pKa = 7.6RR11 pKa = 11.84QRR13 pKa = 11.84AKK15 pKa = 9.44VHH17 pKa = 5.51GFRR20 pKa = 11.84ARR22 pKa = 11.84MEE24 pKa = 4.12TANGRR29 pKa = 11.84KK30 pKa = 9.05VLAARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.87GRR40 pKa = 11.84KK41 pKa = 8.75RR42 pKa = 11.84LAVV45 pKa = 3.41
MM1 pKa = 7.7AKK3 pKa = 8.05MTFQPKK9 pKa = 9.04KK10 pKa = 7.6RR11 pKa = 11.84QRR13 pKa = 11.84AKK15 pKa = 9.44VHH17 pKa = 5.51GFRR20 pKa = 11.84ARR22 pKa = 11.84MEE24 pKa = 4.12TANGRR29 pKa = 11.84KK30 pKa = 9.05VLAARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.87GRR40 pKa = 11.84KK41 pKa = 8.75RR42 pKa = 11.84LAVV45 pKa = 3.41
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
814396 |
39 |
1746 |
347.7 |
38.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.516 ± 0.054 | 1.57 ± 0.019 |
6.497 ± 0.045 | 6.971 ± 0.05 |
4.527 ± 0.039 | 7.03 ± 0.045 |
1.516 ± 0.021 | 7.708 ± 0.043 |
6.648 ± 0.038 | 8.398 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.831 ± 0.023 | 4.584 ± 0.031 |
3.542 ± 0.038 | 2.395 ± 0.021 |
4.031 ± 0.033 | 6.425 ± 0.034 |
5.599 ± 0.041 | 7.071 ± 0.037 |
0.865 ± 0.017 | 4.275 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
