
Halomonas sp. TQ8S
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Halomonas; unclassified Halomonas
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
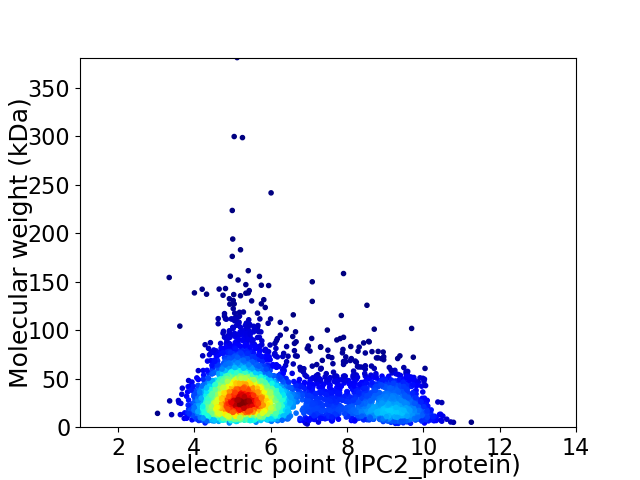
Virtual 2D-PAGE plot for 4010 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A368U602|A0A368U602_9GAMM NAD-dependent protein deacetylase OS=Halomonas sp. TQ8S OX=2282306 GN=DU506_07180 PE=4 SV=1
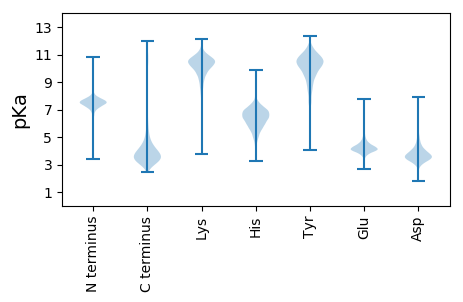
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGAAVMAASGAQAATVYY27 pKa = 11.04NEE29 pKa = 5.19DD30 pKa = 3.42GTKK33 pKa = 10.4LDD35 pKa = 4.25LYY37 pKa = 11.47GNIQIAYY44 pKa = 9.61LNQTQADD51 pKa = 4.02GSDD54 pKa = 3.4EE55 pKa = 6.07DD56 pKa = 3.91EE57 pKa = 5.0LKK59 pKa = 11.28DD60 pKa = 3.5NGSTIGFAGEE70 pKa = 3.88HH71 pKa = 6.43MITRR75 pKa = 11.84DD76 pKa = 3.02LSGYY80 pKa = 9.41FKK82 pKa = 11.41AEE84 pKa = 4.04FEE86 pKa = 4.32HH87 pKa = 7.35DD88 pKa = 3.3ADD90 pKa = 3.95EE91 pKa = 4.67AKK93 pKa = 9.81TAGGLNTGDD102 pKa = 3.17QAYY105 pKa = 9.77IGLKK109 pKa = 10.68GNFGDD114 pKa = 4.2VRR116 pKa = 11.84LGSWDD121 pKa = 3.9PLIDD125 pKa = 5.15DD126 pKa = 5.77WIQDD130 pKa = 3.93PITNNEE136 pKa = 4.08AFDD139 pKa = 3.84VTDD142 pKa = 3.44TNSRR146 pKa = 11.84IVGIEE151 pKa = 3.6NTRR154 pKa = 11.84EE155 pKa = 3.81GDD157 pKa = 3.39KK158 pKa = 10.66LTFTSASLNGLQFAVGTQFEE178 pKa = 4.67GDD180 pKa = 3.67AEE182 pKa = 4.58SDD184 pKa = 3.14GSAVVNPPVGGSLSVSQDD202 pKa = 3.0SGSNASFFGGVKK214 pKa = 8.36YY215 pKa = 8.6TAGAFSVAAVVDD227 pKa = 4.06DD228 pKa = 4.97LGNNDD233 pKa = 3.37GTFTGTDD240 pKa = 3.12ADD242 pKa = 4.25GNTIGGDD249 pKa = 3.46FDD251 pKa = 6.23AGVQYY256 pKa = 11.01GITGQYY262 pKa = 9.02TVDD265 pKa = 3.55TLRR268 pKa = 11.84VALKK272 pKa = 10.6LEE274 pKa = 4.26RR275 pKa = 11.84FEE277 pKa = 5.22SDD279 pKa = 2.44NDD281 pKa = 3.62YY282 pKa = 11.53VADD285 pKa = 3.74TNYY288 pKa = 10.42YY289 pKa = 10.53GLGARR294 pKa = 11.84YY295 pKa = 9.52GYY297 pKa = 10.94GVGDD301 pKa = 3.59IYY303 pKa = 11.28GAYY306 pKa = 9.94QYY308 pKa = 11.63VDD310 pKa = 3.13VGGGNFVDD318 pKa = 4.39TAQDD322 pKa = 3.25AAFSGDD328 pKa = 3.21WPNEE332 pKa = 4.1RR333 pKa = 11.84EE334 pKa = 4.02DD335 pKa = 3.51EE336 pKa = 4.43SYY338 pKa = 11.73NEE340 pKa = 3.95IVLGATYY347 pKa = 10.49NISDD351 pKa = 3.39SMYY354 pKa = 9.79TFIEE358 pKa = 4.21GAMYY362 pKa = 10.79DD363 pKa = 3.62RR364 pKa = 11.84TNDD367 pKa = 3.29EE368 pKa = 4.45DD369 pKa = 5.05DD370 pKa = 4.01GVAVGAVYY378 pKa = 10.44LFF380 pKa = 4.21
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGAAVMAASGAQAATVYY27 pKa = 11.04NEE29 pKa = 5.19DD30 pKa = 3.42GTKK33 pKa = 10.4LDD35 pKa = 4.25LYY37 pKa = 11.47GNIQIAYY44 pKa = 9.61LNQTQADD51 pKa = 4.02GSDD54 pKa = 3.4EE55 pKa = 6.07DD56 pKa = 3.91EE57 pKa = 5.0LKK59 pKa = 11.28DD60 pKa = 3.5NGSTIGFAGEE70 pKa = 3.88HH71 pKa = 6.43MITRR75 pKa = 11.84DD76 pKa = 3.02LSGYY80 pKa = 9.41FKK82 pKa = 11.41AEE84 pKa = 4.04FEE86 pKa = 4.32HH87 pKa = 7.35DD88 pKa = 3.3ADD90 pKa = 3.95EE91 pKa = 4.67AKK93 pKa = 9.81TAGGLNTGDD102 pKa = 3.17QAYY105 pKa = 9.77IGLKK109 pKa = 10.68GNFGDD114 pKa = 4.2VRR116 pKa = 11.84LGSWDD121 pKa = 3.9PLIDD125 pKa = 5.15DD126 pKa = 5.77WIQDD130 pKa = 3.93PITNNEE136 pKa = 4.08AFDD139 pKa = 3.84VTDD142 pKa = 3.44TNSRR146 pKa = 11.84IVGIEE151 pKa = 3.6NTRR154 pKa = 11.84EE155 pKa = 3.81GDD157 pKa = 3.39KK158 pKa = 10.66LTFTSASLNGLQFAVGTQFEE178 pKa = 4.67GDD180 pKa = 3.67AEE182 pKa = 4.58SDD184 pKa = 3.14GSAVVNPPVGGSLSVSQDD202 pKa = 3.0SGSNASFFGGVKK214 pKa = 8.36YY215 pKa = 8.6TAGAFSVAAVVDD227 pKa = 4.06DD228 pKa = 4.97LGNNDD233 pKa = 3.37GTFTGTDD240 pKa = 3.12ADD242 pKa = 4.25GNTIGGDD249 pKa = 3.46FDD251 pKa = 6.23AGVQYY256 pKa = 11.01GITGQYY262 pKa = 9.02TVDD265 pKa = 3.55TLRR268 pKa = 11.84VALKK272 pKa = 10.6LEE274 pKa = 4.26RR275 pKa = 11.84FEE277 pKa = 5.22SDD279 pKa = 2.44NDD281 pKa = 3.62YY282 pKa = 11.53VADD285 pKa = 3.74TNYY288 pKa = 10.42YY289 pKa = 10.53GLGARR294 pKa = 11.84YY295 pKa = 9.52GYY297 pKa = 10.94GVGDD301 pKa = 3.59IYY303 pKa = 11.28GAYY306 pKa = 9.94QYY308 pKa = 11.63VDD310 pKa = 3.13VGGGNFVDD318 pKa = 4.39TAQDD322 pKa = 3.25AAFSGDD328 pKa = 3.21WPNEE332 pKa = 4.1RR333 pKa = 11.84EE334 pKa = 4.02DD335 pKa = 3.51EE336 pKa = 4.43SYY338 pKa = 11.73NEE340 pKa = 3.95IVLGATYY347 pKa = 10.49NISDD351 pKa = 3.39SMYY354 pKa = 9.79TFIEE358 pKa = 4.21GAMYY362 pKa = 10.79DD363 pKa = 3.62RR364 pKa = 11.84TNDD367 pKa = 3.29EE368 pKa = 4.45DD369 pKa = 5.05DD370 pKa = 4.01GVAVGAVYY378 pKa = 10.44LFF380 pKa = 4.21
Molecular weight: 40.24 kDa
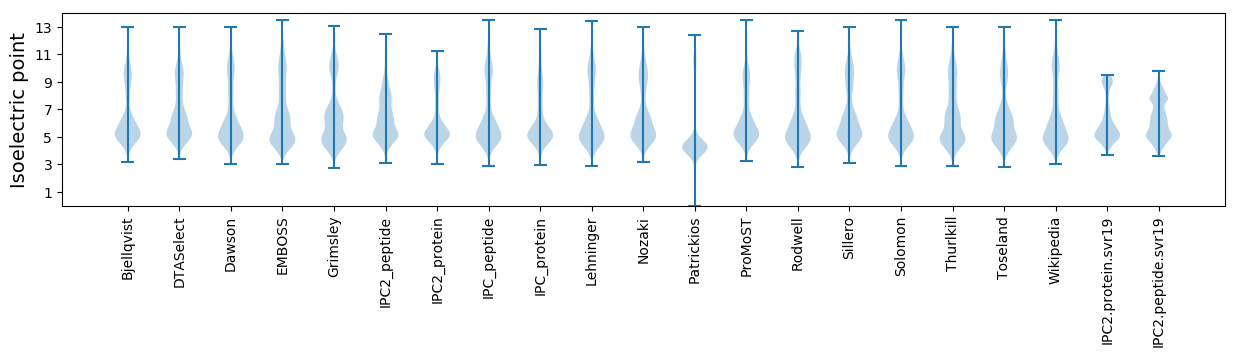
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A368U5U8|A0A368U5U8_9GAMM Histidine kinase OS=Halomonas sp. TQ8S OX=2282306 GN=DU506_07310 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.17NGRR28 pKa = 11.84AILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.17NGRR28 pKa = 11.84AILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1279303 |
30 |
3406 |
319.0 |
35.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.485 ± 0.046 | 0.967 ± 0.014 |
5.591 ± 0.027 | 6.534 ± 0.035 |
3.491 ± 0.024 | 7.622 ± 0.034 |
2.541 ± 0.019 | 4.909 ± 0.025 |
3.334 ± 0.036 | 11.224 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.588 ± 0.021 | 2.908 ± 0.021 |
4.839 ± 0.027 | 4.354 ± 0.032 |
6.809 ± 0.037 | 5.738 ± 0.025 |
5.147 ± 0.026 | 6.972 ± 0.033 |
1.485 ± 0.019 | 2.461 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
