
Aliiglaciecola sp. M165
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Aliiglaciecola; unclassified Aliiglaciecola
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
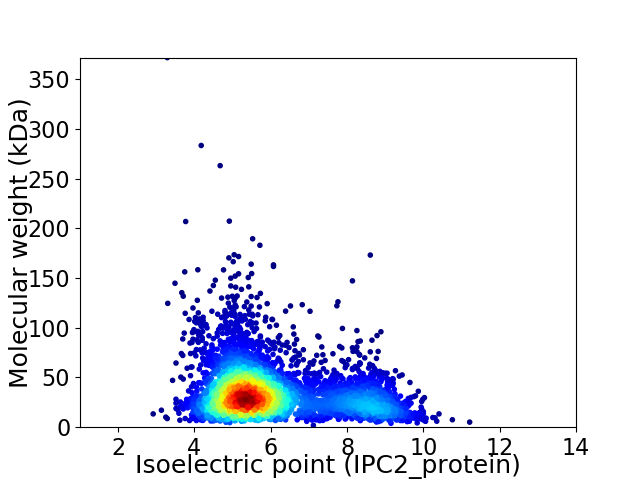
Virtual 2D-PAGE plot for 4070 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A553L388|A0A553L388_9ALTE 30S ribosomal protein S10 OS=Aliiglaciecola sp. M165 OX=2593649 GN=rpsJ PE=3 SV=1
MM1 pKa = 7.65KK2 pKa = 9.13ATSIKK7 pKa = 9.98TPLSLLALALSTAILSGCGSDD28 pKa = 4.59SEE30 pKa = 5.28SIPDD34 pKa = 3.94PVAPPPPPPPPAATDD49 pKa = 4.29FPAQQASPARR59 pKa = 11.84FVDD62 pKa = 3.62GDD64 pKa = 3.6VGEE67 pKa = 4.54VFTTEE72 pKa = 3.76TGLTLYY78 pKa = 10.92YY79 pKa = 10.42FLNDD83 pKa = 3.13EE84 pKa = 4.99AGSSNCNGVEE94 pKa = 3.74GDD96 pKa = 4.23APGSTTDD103 pKa = 3.34EE104 pKa = 4.38TSCAGVWPPLLAATGAQPTGSFSTVEE130 pKa = 4.1RR131 pKa = 11.84DD132 pKa = 4.19DD133 pKa = 4.88GTLQWAYY140 pKa = 11.07RR141 pKa = 11.84DD142 pKa = 3.81FPLYY146 pKa = 10.38TFSNDD151 pKa = 3.03SAQGDD156 pKa = 4.19TNGEE160 pKa = 4.35GINDD164 pKa = 3.31VWFVSRR170 pKa = 11.84PGPSKK175 pKa = 10.34IADD178 pKa = 3.66VNSVPSYY185 pKa = 10.34VGNGTISSVTSTAEE199 pKa = 3.65VLEE202 pKa = 4.18SVRR205 pKa = 11.84LDD207 pKa = 3.29KK208 pKa = 11.39EE209 pKa = 4.43GFSLYY214 pKa = 10.35IFDD217 pKa = 5.97NDD219 pKa = 4.64GLDD222 pKa = 3.28QSACYY227 pKa = 9.84GLNGDD232 pKa = 4.27GCITAWPPLIADD244 pKa = 4.04NAAKK248 pKa = 9.61PAAPLSVVEE257 pKa = 4.64LDD259 pKa = 3.13NGQMQWAYY267 pKa = 10.36RR268 pKa = 11.84GKK270 pKa = 9.45PLYY273 pKa = 10.44FFASDD278 pKa = 3.42TQAGDD283 pKa = 3.5FNGDD287 pKa = 3.01GVGGVWHH294 pKa = 7.25LASQLPAIQRR304 pKa = 11.84EE305 pKa = 4.4VNAQSRR311 pKa = 11.84LSATGRR317 pKa = 11.84VYY319 pKa = 11.29ALLPNTDD326 pKa = 3.69NNNEE330 pKa = 4.16LEE332 pKa = 4.24SQQVDD337 pKa = 2.83KK338 pKa = 11.53DD339 pKa = 3.49QFTVYY344 pKa = 10.37TFDD347 pKa = 3.79NDD349 pKa = 3.63TAGTSNCNGDD359 pKa = 4.72CAVAWPPFLAPDD371 pKa = 4.17AEE373 pKa = 4.19PAVGNFSKK381 pKa = 10.62IEE383 pKa = 4.06RR384 pKa = 11.84TDD386 pKa = 3.88GAIQWAYY393 pKa = 10.72NDD395 pKa = 3.49SPLYY399 pKa = 10.48FFSGDD404 pKa = 3.37NAKK407 pKa = 9.41TDD409 pKa = 3.99LNGDD413 pKa = 3.61GVGGVWHH420 pKa = 6.68IVEE423 pKa = 4.73PPAAPVVSPTSISALSNSLGQTIKK447 pKa = 10.8VDD449 pKa = 3.91GEE451 pKa = 4.22VLALITDD458 pKa = 3.64NQGNSEE464 pKa = 4.4AVTEE468 pKa = 4.37DD469 pKa = 2.97RR470 pKa = 11.84TDD472 pKa = 3.67FQLYY476 pKa = 8.49TFDD479 pKa = 4.15NDD481 pKa = 3.28GDD483 pKa = 4.0EE484 pKa = 5.1LSNCTSTGCMQNWPALLANEE504 pKa = 5.06ADD506 pKa = 3.73TATAPFSIFEE516 pKa = 4.28RR517 pKa = 11.84ADD519 pKa = 3.22GHH521 pKa = 5.76NQWAFNGQPLYY532 pKa = 10.85FFTGDD537 pKa = 3.23TSAGTTNGEE546 pKa = 4.15GVGDD550 pKa = 3.65VWFVARR556 pKa = 11.84PAPVRR561 pKa = 11.84VFNSTDD567 pKa = 3.33DD568 pKa = 3.59GLMIVANDD576 pKa = 4.15LVLASQGKK584 pKa = 6.8TAEE587 pKa = 4.07QLNDD591 pKa = 3.17LTLYY595 pKa = 10.48TFDD598 pKa = 6.18DD599 pKa = 3.84DD600 pKa = 4.25TANSGEE606 pKa = 4.37STCFGGCAVTWPPLYY621 pKa = 9.94ATSADD626 pKa = 3.66QAFGEE631 pKa = 4.38YY632 pKa = 10.12EE633 pKa = 3.93IISRR637 pKa = 11.84TEE639 pKa = 3.7SDD641 pKa = 3.89SSQTLQWTYY650 pKa = 11.33KK651 pKa = 10.45GLPLYY656 pKa = 10.09FFISDD661 pKa = 4.16SEE663 pKa = 4.47LGDD666 pKa = 3.39TGGDD670 pKa = 3.62YY671 pKa = 7.59PTWEE675 pKa = 4.0IARR678 pKa = 11.84PP679 pKa = 3.51
MM1 pKa = 7.65KK2 pKa = 9.13ATSIKK7 pKa = 9.98TPLSLLALALSTAILSGCGSDD28 pKa = 4.59SEE30 pKa = 5.28SIPDD34 pKa = 3.94PVAPPPPPPPPAATDD49 pKa = 4.29FPAQQASPARR59 pKa = 11.84FVDD62 pKa = 3.62GDD64 pKa = 3.6VGEE67 pKa = 4.54VFTTEE72 pKa = 3.76TGLTLYY78 pKa = 10.92YY79 pKa = 10.42FLNDD83 pKa = 3.13EE84 pKa = 4.99AGSSNCNGVEE94 pKa = 3.74GDD96 pKa = 4.23APGSTTDD103 pKa = 3.34EE104 pKa = 4.38TSCAGVWPPLLAATGAQPTGSFSTVEE130 pKa = 4.1RR131 pKa = 11.84DD132 pKa = 4.19DD133 pKa = 4.88GTLQWAYY140 pKa = 11.07RR141 pKa = 11.84DD142 pKa = 3.81FPLYY146 pKa = 10.38TFSNDD151 pKa = 3.03SAQGDD156 pKa = 4.19TNGEE160 pKa = 4.35GINDD164 pKa = 3.31VWFVSRR170 pKa = 11.84PGPSKK175 pKa = 10.34IADD178 pKa = 3.66VNSVPSYY185 pKa = 10.34VGNGTISSVTSTAEE199 pKa = 3.65VLEE202 pKa = 4.18SVRR205 pKa = 11.84LDD207 pKa = 3.29KK208 pKa = 11.39EE209 pKa = 4.43GFSLYY214 pKa = 10.35IFDD217 pKa = 5.97NDD219 pKa = 4.64GLDD222 pKa = 3.28QSACYY227 pKa = 9.84GLNGDD232 pKa = 4.27GCITAWPPLIADD244 pKa = 4.04NAAKK248 pKa = 9.61PAAPLSVVEE257 pKa = 4.64LDD259 pKa = 3.13NGQMQWAYY267 pKa = 10.36RR268 pKa = 11.84GKK270 pKa = 9.45PLYY273 pKa = 10.44FFASDD278 pKa = 3.42TQAGDD283 pKa = 3.5FNGDD287 pKa = 3.01GVGGVWHH294 pKa = 7.25LASQLPAIQRR304 pKa = 11.84EE305 pKa = 4.4VNAQSRR311 pKa = 11.84LSATGRR317 pKa = 11.84VYY319 pKa = 11.29ALLPNTDD326 pKa = 3.69NNNEE330 pKa = 4.16LEE332 pKa = 4.24SQQVDD337 pKa = 2.83KK338 pKa = 11.53DD339 pKa = 3.49QFTVYY344 pKa = 10.37TFDD347 pKa = 3.79NDD349 pKa = 3.63TAGTSNCNGDD359 pKa = 4.72CAVAWPPFLAPDD371 pKa = 4.17AEE373 pKa = 4.19PAVGNFSKK381 pKa = 10.62IEE383 pKa = 4.06RR384 pKa = 11.84TDD386 pKa = 3.88GAIQWAYY393 pKa = 10.72NDD395 pKa = 3.49SPLYY399 pKa = 10.48FFSGDD404 pKa = 3.37NAKK407 pKa = 9.41TDD409 pKa = 3.99LNGDD413 pKa = 3.61GVGGVWHH420 pKa = 6.68IVEE423 pKa = 4.73PPAAPVVSPTSISALSNSLGQTIKK447 pKa = 10.8VDD449 pKa = 3.91GEE451 pKa = 4.22VLALITDD458 pKa = 3.64NQGNSEE464 pKa = 4.4AVTEE468 pKa = 4.37DD469 pKa = 2.97RR470 pKa = 11.84TDD472 pKa = 3.67FQLYY476 pKa = 8.49TFDD479 pKa = 4.15NDD481 pKa = 3.28GDD483 pKa = 4.0EE484 pKa = 5.1LSNCTSTGCMQNWPALLANEE504 pKa = 5.06ADD506 pKa = 3.73TATAPFSIFEE516 pKa = 4.28RR517 pKa = 11.84ADD519 pKa = 3.22GHH521 pKa = 5.76NQWAFNGQPLYY532 pKa = 10.85FFTGDD537 pKa = 3.23TSAGTTNGEE546 pKa = 4.15GVGDD550 pKa = 3.65VWFVARR556 pKa = 11.84PAPVRR561 pKa = 11.84VFNSTDD567 pKa = 3.33DD568 pKa = 3.59GLMIVANDD576 pKa = 4.15LVLASQGKK584 pKa = 6.8TAEE587 pKa = 4.07QLNDD591 pKa = 3.17LTLYY595 pKa = 10.48TFDD598 pKa = 6.18DD599 pKa = 3.84DD600 pKa = 4.25TANSGEE606 pKa = 4.37STCFGGCAVTWPPLYY621 pKa = 9.94ATSADD626 pKa = 3.66QAFGEE631 pKa = 4.38YY632 pKa = 10.12EE633 pKa = 3.93IISRR637 pKa = 11.84TEE639 pKa = 3.7SDD641 pKa = 3.89SSQTLQWTYY650 pKa = 11.33KK651 pKa = 10.45GLPLYY656 pKa = 10.09FFISDD661 pKa = 4.16SEE663 pKa = 4.47LGDD666 pKa = 3.39TGGDD670 pKa = 3.62YY671 pKa = 7.59PTWEE675 pKa = 4.0IARR678 pKa = 11.84PP679 pKa = 3.51
Molecular weight: 72.19 kDa
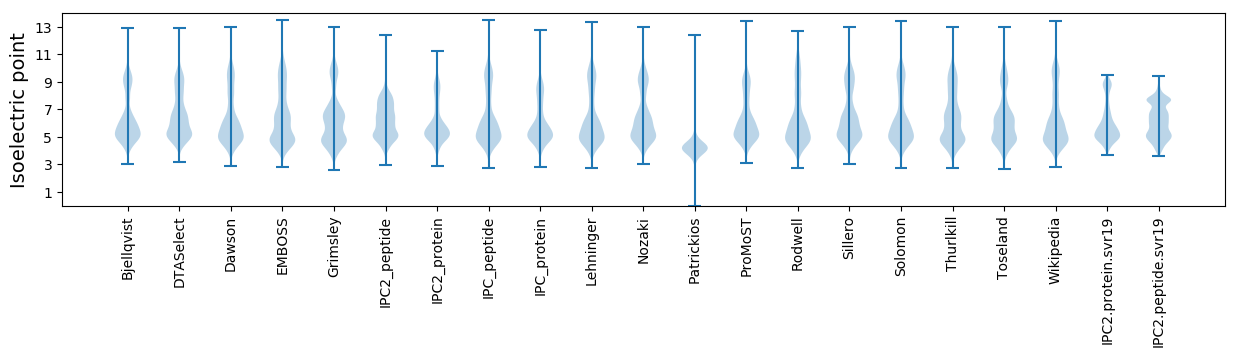
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A553KWG5|A0A553KWG5_9ALTE Monovalent cation/H+ antiporter subunit D OS=Aliiglaciecola sp. M165 OX=2593649 GN=FM019_19940 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.2VLANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.38GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.2VLANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.38GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1360031 |
17 |
3453 |
334.2 |
37.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.319 ± 0.04 | 0.971 ± 0.013 |
5.821 ± 0.035 | 6.155 ± 0.036 |
4.48 ± 0.026 | 6.661 ± 0.039 |
2.219 ± 0.02 | 6.4 ± 0.026 |
5.293 ± 0.038 | 10.193 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.485 ± 0.02 | 4.609 ± 0.028 |
3.857 ± 0.024 | 4.814 ± 0.039 |
4.42 ± 0.027 | 6.852 ± 0.034 |
5.241 ± 0.03 | 6.945 ± 0.031 |
1.257 ± 0.016 | 3.007 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
