
Sclerotinia sclerotiorum mitovirus 6
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.51
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A023J4K6|A0A023J4K6_9VIRU Pol OS=Sclerotinia sclerotiorum mitovirus 6 OX=1435446 PE=4 SV=1
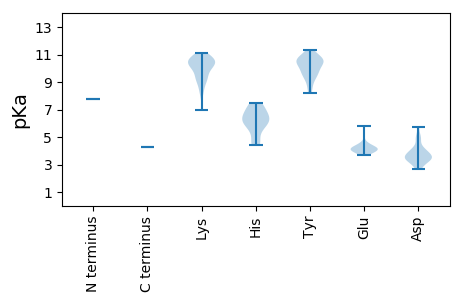
MM1 pKa = 7.76KK2 pKa = 10.23KK3 pKa = 8.1QTYY6 pKa = 9.9KK7 pKa = 10.69LIKK10 pKa = 9.05IICMLFLNIGRR21 pKa = 11.84HH22 pKa = 4.56TYY24 pKa = 10.71VIGLVMKK31 pKa = 10.5RR32 pKa = 11.84IHH34 pKa = 6.72NLLKK38 pKa = 11.15NNGTLFTVKK47 pKa = 10.21YY48 pKa = 9.35LKK50 pKa = 9.76QARR53 pKa = 11.84LHH55 pKa = 4.39VTRR58 pKa = 11.84YY59 pKa = 9.1YY60 pKa = 10.67CGSPLMEE67 pKa = 4.48NNIGVSLTQTGFPIIFKK84 pKa = 10.12EE85 pKa = 4.15CEE87 pKa = 3.84PLIKK91 pKa = 10.65GSMNLKK97 pKa = 10.18KK98 pKa = 10.63FVFTLINISRR108 pKa = 11.84TIRR111 pKa = 11.84PRR113 pKa = 11.84KK114 pKa = 10.09GEE116 pKa = 4.34IIPISLKK123 pKa = 10.66PITDD127 pKa = 3.67PSTAVINKK135 pKa = 9.5LDD137 pKa = 3.5NRR139 pKa = 11.84LLRR142 pKa = 11.84KK143 pKa = 9.93VISQNDD149 pKa = 3.31ICLSIPDD156 pKa = 4.74HH157 pKa = 6.25GLSDD161 pKa = 4.92LEE163 pKa = 4.48LTTKK167 pKa = 10.41SGPHH171 pKa = 6.31GPQTLTALYY180 pKa = 10.7SMKK183 pKa = 10.28RR184 pKa = 11.84YY185 pKa = 8.24STATASAIMGMLSTSLFKK203 pKa = 11.0YY204 pKa = 10.5FKK206 pKa = 10.57DD207 pKa = 3.67LYY209 pKa = 10.37QYY211 pKa = 11.3SLSKK215 pKa = 10.14PDD217 pKa = 3.36EE218 pKa = 4.23VKK220 pKa = 10.78PCGCKK225 pKa = 9.68AAPEE229 pKa = 4.0VRR231 pKa = 11.84RR232 pKa = 11.84LSIVKK237 pKa = 10.3DD238 pKa = 3.94PEE240 pKa = 3.84CKK242 pKa = 9.42MRR244 pKa = 11.84VIGMFDD250 pKa = 2.67WWSQVTLKK258 pKa = 10.49PLSDD262 pKa = 4.07SLFKK266 pKa = 10.9ALSKK270 pKa = 10.17IDD272 pKa = 3.34SDD274 pKa = 3.51RR275 pKa = 11.84TYY277 pKa = 10.72SQDD280 pKa = 3.36PAFHH284 pKa = 6.92FEE286 pKa = 3.91VDD288 pKa = 3.83TNLLWSIDD296 pKa = 3.47LTAATDD302 pKa = 3.81RR303 pKa = 11.84FPIEE307 pKa = 3.95LQKK310 pKa = 10.99QILSILTNTAFTRR323 pKa = 11.84DD324 pKa = 3.32WADD327 pKa = 3.0VMVGEE332 pKa = 4.36EE333 pKa = 3.97FSYY336 pKa = 11.01EE337 pKa = 4.02GGSISYY343 pKa = 10.22RR344 pKa = 11.84VGQPMGAYY352 pKa = 9.93SSWPMFTLSHH362 pKa = 6.87HH363 pKa = 5.92ILVRR367 pKa = 11.84YY368 pKa = 9.74CGLLNGIDD376 pKa = 3.5NFNRR380 pKa = 11.84YY381 pKa = 8.65IMLGDD386 pKa = 5.05DD387 pKa = 4.19IVINHH392 pKa = 6.18NKK394 pKa = 8.37VAKK397 pKa = 9.28TYY399 pKa = 10.38IRR401 pKa = 11.84LLHH404 pKa = 6.22ALGVEE409 pKa = 4.66TSQAKK414 pKa = 6.99THH416 pKa = 5.39VSKK419 pKa = 9.2HH420 pKa = 4.6TYY422 pKa = 9.7EE423 pKa = 4.18FAKK426 pKa = 10.17RR427 pKa = 11.84WIDD430 pKa = 3.51TRR432 pKa = 11.84CGEE435 pKa = 4.16ITGLPIKK442 pKa = 10.62GLLDD446 pKa = 4.13NIEE449 pKa = 4.08NMSTCFQILFDD460 pKa = 4.03WQLKK464 pKa = 8.97GNKK467 pKa = 8.7FWTDD471 pKa = 2.83RR472 pKa = 11.84SLVEE476 pKa = 4.17SVADD480 pKa = 3.63FLYY483 pKa = 10.53KK484 pKa = 10.69VQDD487 pKa = 4.21FSKK490 pKa = 10.7KK491 pKa = 8.34PYY493 pKa = 9.57GYY495 pKa = 10.48RR496 pKa = 11.84KK497 pKa = 9.4ILEE500 pKa = 4.19TLKK503 pKa = 10.64PISFFMRR510 pKa = 11.84LRR512 pKa = 11.84FSLSTPDD519 pKa = 3.17EE520 pKa = 4.17VRR522 pKa = 11.84NQICLWTRR530 pKa = 11.84NSQFIVGGAEE540 pKa = 3.78KK541 pKa = 10.76LILNEE546 pKa = 3.72VLRR549 pKa = 11.84VFNLTYY555 pKa = 10.41SALVQKK561 pKa = 10.35SSRR564 pKa = 11.84LIVEE568 pKa = 5.18FYY570 pKa = 10.79HH571 pKa = 7.33RR572 pKa = 11.84INEE575 pKa = 4.21FVEE578 pKa = 4.5LEE580 pKa = 4.01DD581 pKa = 5.7NIIEE585 pKa = 4.32DD586 pKa = 3.96HH587 pKa = 5.8VHH589 pKa = 7.46PILQCIEE596 pKa = 3.83NHH598 pKa = 6.19LEE600 pKa = 4.24RR601 pKa = 11.84IAEE604 pKa = 4.12KK605 pKa = 10.63QEE607 pKa = 4.0SQRR610 pKa = 11.84GDD612 pKa = 3.27LTLKK616 pKa = 10.12EE617 pKa = 4.16MCDD620 pKa = 3.6TLMLMDD626 pKa = 4.98IEE628 pKa = 5.82SIMTQKK634 pKa = 8.98RR635 pKa = 11.84TKK637 pKa = 9.24VQIMVNNTKK646 pKa = 9.36MAHH649 pKa = 6.01KK650 pKa = 9.87FVNLVKK656 pKa = 10.52SWDD659 pKa = 3.47HH660 pKa = 6.62DD661 pKa = 3.72NPRR664 pKa = 11.84FSDD667 pKa = 3.6PQNFFHH673 pKa = 7.3GGAADD678 pKa = 3.21IGFGSTSVQQTLDD691 pKa = 2.67RR692 pKa = 11.84FRR694 pKa = 11.84RR695 pKa = 11.84YY696 pKa = 9.47FLL698 pKa = 4.27
MM1 pKa = 7.76KK2 pKa = 10.23KK3 pKa = 8.1QTYY6 pKa = 9.9KK7 pKa = 10.69LIKK10 pKa = 9.05IICMLFLNIGRR21 pKa = 11.84HH22 pKa = 4.56TYY24 pKa = 10.71VIGLVMKK31 pKa = 10.5RR32 pKa = 11.84IHH34 pKa = 6.72NLLKK38 pKa = 11.15NNGTLFTVKK47 pKa = 10.21YY48 pKa = 9.35LKK50 pKa = 9.76QARR53 pKa = 11.84LHH55 pKa = 4.39VTRR58 pKa = 11.84YY59 pKa = 9.1YY60 pKa = 10.67CGSPLMEE67 pKa = 4.48NNIGVSLTQTGFPIIFKK84 pKa = 10.12EE85 pKa = 4.15CEE87 pKa = 3.84PLIKK91 pKa = 10.65GSMNLKK97 pKa = 10.18KK98 pKa = 10.63FVFTLINISRR108 pKa = 11.84TIRR111 pKa = 11.84PRR113 pKa = 11.84KK114 pKa = 10.09GEE116 pKa = 4.34IIPISLKK123 pKa = 10.66PITDD127 pKa = 3.67PSTAVINKK135 pKa = 9.5LDD137 pKa = 3.5NRR139 pKa = 11.84LLRR142 pKa = 11.84KK143 pKa = 9.93VISQNDD149 pKa = 3.31ICLSIPDD156 pKa = 4.74HH157 pKa = 6.25GLSDD161 pKa = 4.92LEE163 pKa = 4.48LTTKK167 pKa = 10.41SGPHH171 pKa = 6.31GPQTLTALYY180 pKa = 10.7SMKK183 pKa = 10.28RR184 pKa = 11.84YY185 pKa = 8.24STATASAIMGMLSTSLFKK203 pKa = 11.0YY204 pKa = 10.5FKK206 pKa = 10.57DD207 pKa = 3.67LYY209 pKa = 10.37QYY211 pKa = 11.3SLSKK215 pKa = 10.14PDD217 pKa = 3.36EE218 pKa = 4.23VKK220 pKa = 10.78PCGCKK225 pKa = 9.68AAPEE229 pKa = 4.0VRR231 pKa = 11.84RR232 pKa = 11.84LSIVKK237 pKa = 10.3DD238 pKa = 3.94PEE240 pKa = 3.84CKK242 pKa = 9.42MRR244 pKa = 11.84VIGMFDD250 pKa = 2.67WWSQVTLKK258 pKa = 10.49PLSDD262 pKa = 4.07SLFKK266 pKa = 10.9ALSKK270 pKa = 10.17IDD272 pKa = 3.34SDD274 pKa = 3.51RR275 pKa = 11.84TYY277 pKa = 10.72SQDD280 pKa = 3.36PAFHH284 pKa = 6.92FEE286 pKa = 3.91VDD288 pKa = 3.83TNLLWSIDD296 pKa = 3.47LTAATDD302 pKa = 3.81RR303 pKa = 11.84FPIEE307 pKa = 3.95LQKK310 pKa = 10.99QILSILTNTAFTRR323 pKa = 11.84DD324 pKa = 3.32WADD327 pKa = 3.0VMVGEE332 pKa = 4.36EE333 pKa = 3.97FSYY336 pKa = 11.01EE337 pKa = 4.02GGSISYY343 pKa = 10.22RR344 pKa = 11.84VGQPMGAYY352 pKa = 9.93SSWPMFTLSHH362 pKa = 6.87HH363 pKa = 5.92ILVRR367 pKa = 11.84YY368 pKa = 9.74CGLLNGIDD376 pKa = 3.5NFNRR380 pKa = 11.84YY381 pKa = 8.65IMLGDD386 pKa = 5.05DD387 pKa = 4.19IVINHH392 pKa = 6.18NKK394 pKa = 8.37VAKK397 pKa = 9.28TYY399 pKa = 10.38IRR401 pKa = 11.84LLHH404 pKa = 6.22ALGVEE409 pKa = 4.66TSQAKK414 pKa = 6.99THH416 pKa = 5.39VSKK419 pKa = 9.2HH420 pKa = 4.6TYY422 pKa = 9.7EE423 pKa = 4.18FAKK426 pKa = 10.17RR427 pKa = 11.84WIDD430 pKa = 3.51TRR432 pKa = 11.84CGEE435 pKa = 4.16ITGLPIKK442 pKa = 10.62GLLDD446 pKa = 4.13NIEE449 pKa = 4.08NMSTCFQILFDD460 pKa = 4.03WQLKK464 pKa = 8.97GNKK467 pKa = 8.7FWTDD471 pKa = 2.83RR472 pKa = 11.84SLVEE476 pKa = 4.17SVADD480 pKa = 3.63FLYY483 pKa = 10.53KK484 pKa = 10.69VQDD487 pKa = 4.21FSKK490 pKa = 10.7KK491 pKa = 8.34PYY493 pKa = 9.57GYY495 pKa = 10.48RR496 pKa = 11.84KK497 pKa = 9.4ILEE500 pKa = 4.19TLKK503 pKa = 10.64PISFFMRR510 pKa = 11.84LRR512 pKa = 11.84FSLSTPDD519 pKa = 3.17EE520 pKa = 4.17VRR522 pKa = 11.84NQICLWTRR530 pKa = 11.84NSQFIVGGAEE540 pKa = 3.78KK541 pKa = 10.76LILNEE546 pKa = 3.72VLRR549 pKa = 11.84VFNLTYY555 pKa = 10.41SALVQKK561 pKa = 10.35SSRR564 pKa = 11.84LIVEE568 pKa = 5.18FYY570 pKa = 10.79HH571 pKa = 7.33RR572 pKa = 11.84INEE575 pKa = 4.21FVEE578 pKa = 4.5LEE580 pKa = 4.01DD581 pKa = 5.7NIIEE585 pKa = 4.32DD586 pKa = 3.96HH587 pKa = 5.8VHH589 pKa = 7.46PILQCIEE596 pKa = 3.83NHH598 pKa = 6.19LEE600 pKa = 4.24RR601 pKa = 11.84IAEE604 pKa = 4.12KK605 pKa = 10.63QEE607 pKa = 4.0SQRR610 pKa = 11.84GDD612 pKa = 3.27LTLKK616 pKa = 10.12EE617 pKa = 4.16MCDD620 pKa = 3.6TLMLMDD626 pKa = 4.98IEE628 pKa = 5.82SIMTQKK634 pKa = 8.98RR635 pKa = 11.84TKK637 pKa = 9.24VQIMVNNTKK646 pKa = 9.36MAHH649 pKa = 6.01KK650 pKa = 9.87FVNLVKK656 pKa = 10.52SWDD659 pKa = 3.47HH660 pKa = 6.62DD661 pKa = 3.72NPRR664 pKa = 11.84FSDD667 pKa = 3.6PQNFFHH673 pKa = 7.3GGAADD678 pKa = 3.21IGFGSTSVQQTLDD691 pKa = 2.67RR692 pKa = 11.84FRR694 pKa = 11.84RR695 pKa = 11.84YY696 pKa = 9.47FLL698 pKa = 4.27
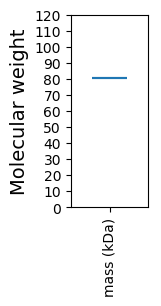
Molecular weight: 80.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A023J4K6|A0A023J4K6_9VIRU Pol OS=Sclerotinia sclerotiorum mitovirus 6 OX=1435446 PE=4 SV=1
MM1 pKa = 7.76KK2 pKa = 10.23KK3 pKa = 8.1QTYY6 pKa = 9.9KK7 pKa = 10.69LIKK10 pKa = 9.05IICMLFLNIGRR21 pKa = 11.84HH22 pKa = 4.56TYY24 pKa = 10.71VIGLVMKK31 pKa = 10.5RR32 pKa = 11.84IHH34 pKa = 6.72NLLKK38 pKa = 11.15NNGTLFTVKK47 pKa = 10.21YY48 pKa = 9.35LKK50 pKa = 9.76QARR53 pKa = 11.84LHH55 pKa = 4.39VTRR58 pKa = 11.84YY59 pKa = 9.1YY60 pKa = 10.67CGSPLMEE67 pKa = 4.48NNIGVSLTQTGFPIIFKK84 pKa = 10.12EE85 pKa = 4.15CEE87 pKa = 3.84PLIKK91 pKa = 10.65GSMNLKK97 pKa = 10.18KK98 pKa = 10.63FVFTLINISRR108 pKa = 11.84TIRR111 pKa = 11.84PRR113 pKa = 11.84KK114 pKa = 10.09GEE116 pKa = 4.34IIPISLKK123 pKa = 10.66PITDD127 pKa = 3.67PSTAVINKK135 pKa = 9.5LDD137 pKa = 3.5NRR139 pKa = 11.84LLRR142 pKa = 11.84KK143 pKa = 9.93VISQNDD149 pKa = 3.31ICLSIPDD156 pKa = 4.74HH157 pKa = 6.25GLSDD161 pKa = 4.92LEE163 pKa = 4.48LTTKK167 pKa = 10.41SGPHH171 pKa = 6.31GPQTLTALYY180 pKa = 10.7SMKK183 pKa = 10.28RR184 pKa = 11.84YY185 pKa = 8.24STATASAIMGMLSTSLFKK203 pKa = 11.0YY204 pKa = 10.5FKK206 pKa = 10.57DD207 pKa = 3.67LYY209 pKa = 10.37QYY211 pKa = 11.3SLSKK215 pKa = 10.14PDD217 pKa = 3.36EE218 pKa = 4.23VKK220 pKa = 10.78PCGCKK225 pKa = 9.68AAPEE229 pKa = 4.0VRR231 pKa = 11.84RR232 pKa = 11.84LSIVKK237 pKa = 10.3DD238 pKa = 3.94PEE240 pKa = 3.84CKK242 pKa = 9.42MRR244 pKa = 11.84VIGMFDD250 pKa = 2.67WWSQVTLKK258 pKa = 10.49PLSDD262 pKa = 4.07SLFKK266 pKa = 10.9ALSKK270 pKa = 10.17IDD272 pKa = 3.34SDD274 pKa = 3.51RR275 pKa = 11.84TYY277 pKa = 10.72SQDD280 pKa = 3.36PAFHH284 pKa = 6.92FEE286 pKa = 3.91VDD288 pKa = 3.83TNLLWSIDD296 pKa = 3.47LTAATDD302 pKa = 3.81RR303 pKa = 11.84FPIEE307 pKa = 3.95LQKK310 pKa = 10.99QILSILTNTAFTRR323 pKa = 11.84DD324 pKa = 3.32WADD327 pKa = 3.0VMVGEE332 pKa = 4.36EE333 pKa = 3.97FSYY336 pKa = 11.01EE337 pKa = 4.02GGSISYY343 pKa = 10.22RR344 pKa = 11.84VGQPMGAYY352 pKa = 9.93SSWPMFTLSHH362 pKa = 6.87HH363 pKa = 5.92ILVRR367 pKa = 11.84YY368 pKa = 9.74CGLLNGIDD376 pKa = 3.5NFNRR380 pKa = 11.84YY381 pKa = 8.65IMLGDD386 pKa = 5.05DD387 pKa = 4.19IVINHH392 pKa = 6.18NKK394 pKa = 8.37VAKK397 pKa = 9.28TYY399 pKa = 10.38IRR401 pKa = 11.84LLHH404 pKa = 6.22ALGVEE409 pKa = 4.66TSQAKK414 pKa = 6.99THH416 pKa = 5.39VSKK419 pKa = 9.2HH420 pKa = 4.6TYY422 pKa = 9.7EE423 pKa = 4.18FAKK426 pKa = 10.17RR427 pKa = 11.84WIDD430 pKa = 3.51TRR432 pKa = 11.84CGEE435 pKa = 4.16ITGLPIKK442 pKa = 10.62GLLDD446 pKa = 4.13NIEE449 pKa = 4.08NMSTCFQILFDD460 pKa = 4.03WQLKK464 pKa = 8.97GNKK467 pKa = 8.7FWTDD471 pKa = 2.83RR472 pKa = 11.84SLVEE476 pKa = 4.17SVADD480 pKa = 3.63FLYY483 pKa = 10.53KK484 pKa = 10.69VQDD487 pKa = 4.21FSKK490 pKa = 10.7KK491 pKa = 8.34PYY493 pKa = 9.57GYY495 pKa = 10.48RR496 pKa = 11.84KK497 pKa = 9.4ILEE500 pKa = 4.19TLKK503 pKa = 10.64PISFFMRR510 pKa = 11.84LRR512 pKa = 11.84FSLSTPDD519 pKa = 3.17EE520 pKa = 4.17VRR522 pKa = 11.84NQICLWTRR530 pKa = 11.84NSQFIVGGAEE540 pKa = 3.78KK541 pKa = 10.76LILNEE546 pKa = 3.72VLRR549 pKa = 11.84VFNLTYY555 pKa = 10.41SALVQKK561 pKa = 10.35SSRR564 pKa = 11.84LIVEE568 pKa = 5.18FYY570 pKa = 10.79HH571 pKa = 7.33RR572 pKa = 11.84INEE575 pKa = 4.21FVEE578 pKa = 4.5LEE580 pKa = 4.01DD581 pKa = 5.7NIIEE585 pKa = 4.32DD586 pKa = 3.96HH587 pKa = 5.8VHH589 pKa = 7.46PILQCIEE596 pKa = 3.83NHH598 pKa = 6.19LEE600 pKa = 4.24RR601 pKa = 11.84IAEE604 pKa = 4.12KK605 pKa = 10.63QEE607 pKa = 4.0SQRR610 pKa = 11.84GDD612 pKa = 3.27LTLKK616 pKa = 10.12EE617 pKa = 4.16MCDD620 pKa = 3.6TLMLMDD626 pKa = 4.98IEE628 pKa = 5.82SIMTQKK634 pKa = 8.98RR635 pKa = 11.84TKK637 pKa = 9.24VQIMVNNTKK646 pKa = 9.36MAHH649 pKa = 6.01KK650 pKa = 9.87FVNLVKK656 pKa = 10.52SWDD659 pKa = 3.47HH660 pKa = 6.62DD661 pKa = 3.72NPRR664 pKa = 11.84FSDD667 pKa = 3.6PQNFFHH673 pKa = 7.3GGAADD678 pKa = 3.21IGFGSTSVQQTLDD691 pKa = 2.67RR692 pKa = 11.84FRR694 pKa = 11.84RR695 pKa = 11.84YY696 pKa = 9.47FLL698 pKa = 4.27
MM1 pKa = 7.76KK2 pKa = 10.23KK3 pKa = 8.1QTYY6 pKa = 9.9KK7 pKa = 10.69LIKK10 pKa = 9.05IICMLFLNIGRR21 pKa = 11.84HH22 pKa = 4.56TYY24 pKa = 10.71VIGLVMKK31 pKa = 10.5RR32 pKa = 11.84IHH34 pKa = 6.72NLLKK38 pKa = 11.15NNGTLFTVKK47 pKa = 10.21YY48 pKa = 9.35LKK50 pKa = 9.76QARR53 pKa = 11.84LHH55 pKa = 4.39VTRR58 pKa = 11.84YY59 pKa = 9.1YY60 pKa = 10.67CGSPLMEE67 pKa = 4.48NNIGVSLTQTGFPIIFKK84 pKa = 10.12EE85 pKa = 4.15CEE87 pKa = 3.84PLIKK91 pKa = 10.65GSMNLKK97 pKa = 10.18KK98 pKa = 10.63FVFTLINISRR108 pKa = 11.84TIRR111 pKa = 11.84PRR113 pKa = 11.84KK114 pKa = 10.09GEE116 pKa = 4.34IIPISLKK123 pKa = 10.66PITDD127 pKa = 3.67PSTAVINKK135 pKa = 9.5LDD137 pKa = 3.5NRR139 pKa = 11.84LLRR142 pKa = 11.84KK143 pKa = 9.93VISQNDD149 pKa = 3.31ICLSIPDD156 pKa = 4.74HH157 pKa = 6.25GLSDD161 pKa = 4.92LEE163 pKa = 4.48LTTKK167 pKa = 10.41SGPHH171 pKa = 6.31GPQTLTALYY180 pKa = 10.7SMKK183 pKa = 10.28RR184 pKa = 11.84YY185 pKa = 8.24STATASAIMGMLSTSLFKK203 pKa = 11.0YY204 pKa = 10.5FKK206 pKa = 10.57DD207 pKa = 3.67LYY209 pKa = 10.37QYY211 pKa = 11.3SLSKK215 pKa = 10.14PDD217 pKa = 3.36EE218 pKa = 4.23VKK220 pKa = 10.78PCGCKK225 pKa = 9.68AAPEE229 pKa = 4.0VRR231 pKa = 11.84RR232 pKa = 11.84LSIVKK237 pKa = 10.3DD238 pKa = 3.94PEE240 pKa = 3.84CKK242 pKa = 9.42MRR244 pKa = 11.84VIGMFDD250 pKa = 2.67WWSQVTLKK258 pKa = 10.49PLSDD262 pKa = 4.07SLFKK266 pKa = 10.9ALSKK270 pKa = 10.17IDD272 pKa = 3.34SDD274 pKa = 3.51RR275 pKa = 11.84TYY277 pKa = 10.72SQDD280 pKa = 3.36PAFHH284 pKa = 6.92FEE286 pKa = 3.91VDD288 pKa = 3.83TNLLWSIDD296 pKa = 3.47LTAATDD302 pKa = 3.81RR303 pKa = 11.84FPIEE307 pKa = 3.95LQKK310 pKa = 10.99QILSILTNTAFTRR323 pKa = 11.84DD324 pKa = 3.32WADD327 pKa = 3.0VMVGEE332 pKa = 4.36EE333 pKa = 3.97FSYY336 pKa = 11.01EE337 pKa = 4.02GGSISYY343 pKa = 10.22RR344 pKa = 11.84VGQPMGAYY352 pKa = 9.93SSWPMFTLSHH362 pKa = 6.87HH363 pKa = 5.92ILVRR367 pKa = 11.84YY368 pKa = 9.74CGLLNGIDD376 pKa = 3.5NFNRR380 pKa = 11.84YY381 pKa = 8.65IMLGDD386 pKa = 5.05DD387 pKa = 4.19IVINHH392 pKa = 6.18NKK394 pKa = 8.37VAKK397 pKa = 9.28TYY399 pKa = 10.38IRR401 pKa = 11.84LLHH404 pKa = 6.22ALGVEE409 pKa = 4.66TSQAKK414 pKa = 6.99THH416 pKa = 5.39VSKK419 pKa = 9.2HH420 pKa = 4.6TYY422 pKa = 9.7EE423 pKa = 4.18FAKK426 pKa = 10.17RR427 pKa = 11.84WIDD430 pKa = 3.51TRR432 pKa = 11.84CGEE435 pKa = 4.16ITGLPIKK442 pKa = 10.62GLLDD446 pKa = 4.13NIEE449 pKa = 4.08NMSTCFQILFDD460 pKa = 4.03WQLKK464 pKa = 8.97GNKK467 pKa = 8.7FWTDD471 pKa = 2.83RR472 pKa = 11.84SLVEE476 pKa = 4.17SVADD480 pKa = 3.63FLYY483 pKa = 10.53KK484 pKa = 10.69VQDD487 pKa = 4.21FSKK490 pKa = 10.7KK491 pKa = 8.34PYY493 pKa = 9.57GYY495 pKa = 10.48RR496 pKa = 11.84KK497 pKa = 9.4ILEE500 pKa = 4.19TLKK503 pKa = 10.64PISFFMRR510 pKa = 11.84LRR512 pKa = 11.84FSLSTPDD519 pKa = 3.17EE520 pKa = 4.17VRR522 pKa = 11.84NQICLWTRR530 pKa = 11.84NSQFIVGGAEE540 pKa = 3.78KK541 pKa = 10.76LILNEE546 pKa = 3.72VLRR549 pKa = 11.84VFNLTYY555 pKa = 10.41SALVQKK561 pKa = 10.35SSRR564 pKa = 11.84LIVEE568 pKa = 5.18FYY570 pKa = 10.79HH571 pKa = 7.33RR572 pKa = 11.84INEE575 pKa = 4.21FVEE578 pKa = 4.5LEE580 pKa = 4.01DD581 pKa = 5.7NIIEE585 pKa = 4.32DD586 pKa = 3.96HH587 pKa = 5.8VHH589 pKa = 7.46PILQCIEE596 pKa = 3.83NHH598 pKa = 6.19LEE600 pKa = 4.24RR601 pKa = 11.84IAEE604 pKa = 4.12KK605 pKa = 10.63QEE607 pKa = 4.0SQRR610 pKa = 11.84GDD612 pKa = 3.27LTLKK616 pKa = 10.12EE617 pKa = 4.16MCDD620 pKa = 3.6TLMLMDD626 pKa = 4.98IEE628 pKa = 5.82SIMTQKK634 pKa = 8.98RR635 pKa = 11.84TKK637 pKa = 9.24VQIMVNNTKK646 pKa = 9.36MAHH649 pKa = 6.01KK650 pKa = 9.87FVNLVKK656 pKa = 10.52SWDD659 pKa = 3.47HH660 pKa = 6.62DD661 pKa = 3.72NPRR664 pKa = 11.84FSDD667 pKa = 3.6PQNFFHH673 pKa = 7.3GGAADD678 pKa = 3.21IGFGSTSVQQTLDD691 pKa = 2.67RR692 pKa = 11.84FRR694 pKa = 11.84RR695 pKa = 11.84YY696 pKa = 9.47FLL698 pKa = 4.27
Molecular weight: 80.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
698 |
698 |
698 |
698.0 |
80.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.725 ± 0.0 | 1.862 ± 0.0 |
5.444 ± 0.0 | 4.728 ± 0.0 |
5.301 ± 0.0 | 5.014 ± 0.0 |
2.722 ± 0.0 | 8.166 ± 0.0 |
7.307 ± 0.0 | 10.745 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.152 ± 0.0 | 4.728 ± 0.0 |
3.725 ± 0.0 | 3.725 ± 0.0 |
5.444 ± 0.0 | 7.307 ± 0.0 |
6.59 ± 0.0 | 5.444 ± 0.0 |
1.433 ± 0.0 | 3.438 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
