
Azoarcus pumilus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Rhodocyclales; Zoogloeaceae; Azoarcus
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
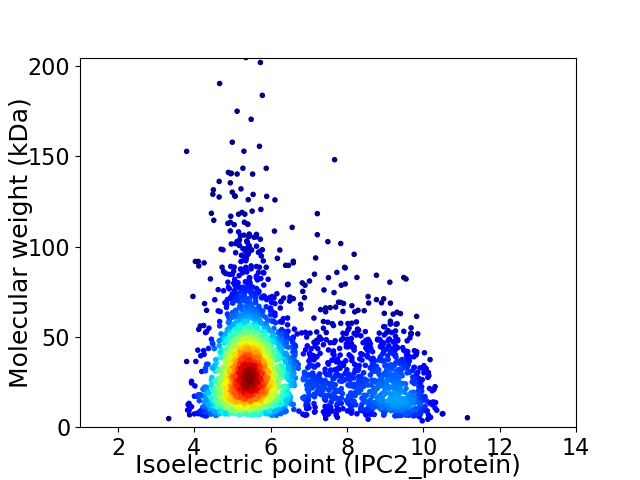
Virtual 2D-PAGE plot for 3041 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I6S3A3|A0A2I6S3A3_9RHOO FecR domain-containing protein OS=Azoarcus pumilus OX=2067960 GN=C0099_01545 PE=4 SV=1
MM1 pKa = 7.98IGIEE5 pKa = 4.22LLQGAARR12 pKa = 11.84RR13 pKa = 11.84EE14 pKa = 4.24SGRR17 pKa = 11.84GVNARR22 pKa = 11.84SAARR26 pKa = 11.84RR27 pKa = 11.84LLAVLRR33 pKa = 11.84GLIFVPGLVLAAGTAMAQNVDD54 pKa = 3.57LVLNHH59 pKa = 7.33DD60 pKa = 4.19LTSAATINASDD71 pKa = 3.53TAAFTVTVDD80 pKa = 3.75NSDD83 pKa = 3.5PTHH86 pKa = 6.57TGDD89 pKa = 3.5ANNVVVTYY97 pKa = 9.91QISATDD103 pKa = 3.85TLLSASPSQGSCSLPPDD120 pKa = 3.59ASNTFTCNFGTIPHH134 pKa = 5.84TTNATLDD141 pKa = 3.67VVVRR145 pKa = 11.84ATGTGVGSFPYY156 pKa = 10.73TMTTNAAVATSSTDD170 pKa = 3.35TEE172 pKa = 4.1ATNNDD177 pKa = 3.37EE178 pKa = 4.22EE179 pKa = 4.57RR180 pKa = 11.84TVTVQEE186 pKa = 4.33GTDD189 pKa = 3.29VGIVLNSSPGAIASGGIWTHH209 pKa = 5.33EE210 pKa = 3.84MVVTNHH216 pKa = 5.72GPIGATNIVAHH227 pKa = 7.06LDD229 pKa = 3.73LPAGAILVGSLPAGCSAGAPGVVCQIASLALSGVQNIGPITTQIAATSGSNLSATALVDD288 pKa = 5.08SLDD291 pKa = 3.72QYY293 pKa = 11.88DD294 pKa = 4.51GNTTNQSSTRR304 pKa = 11.84QVDD307 pKa = 3.67VNPGSDD313 pKa = 3.26LAVTLGQSGGPFLEE327 pKa = 4.4NEE329 pKa = 4.25PFALTANPSYY339 pKa = 10.17TGEE342 pKa = 4.2VPDD345 pKa = 4.19VPEE348 pKa = 4.64LSISVDD354 pKa = 3.08PGLQIQNPAVFNQNGWTCSVSGQLVSCSGLDD385 pKa = 3.57TSGMTAGANQPLGVVTVNVVGDD407 pKa = 3.53TAGTYY412 pKa = 9.48SANEE416 pKa = 3.94ATVSADD422 pKa = 3.44YY423 pKa = 10.54TNKK426 pKa = 9.74PDD428 pKa = 4.37PNLDD432 pKa = 3.54NNIDD436 pKa = 3.6GTAPAIVAAEE446 pKa = 4.1LNLGIVKK453 pKa = 10.14SAPQRR458 pKa = 11.84DD459 pKa = 3.91LRR461 pKa = 11.84TIGAAYY467 pKa = 9.18PFDD470 pKa = 3.94YY471 pKa = 10.89SVTVRR476 pKa = 11.84NNGNVGFWGDD486 pKa = 3.93LTWTDD491 pKa = 3.94TVPEE495 pKa = 4.58GLTITNIAAPGGWTCTTVPPNVVGADD521 pKa = 3.22TFTCSRR527 pKa = 11.84TYY529 pKa = 11.43AEE531 pKa = 4.52GAPFAVNATQAFTITAFANGDD552 pKa = 3.41IVGDD556 pKa = 4.08LVNGACLTTIDD567 pKa = 4.43HH568 pKa = 7.51DD569 pKa = 4.86LGFLEE574 pKa = 5.28NEE576 pKa = 4.26VATPCSNATINAQTPGSSADD596 pKa = 3.7LLVEE600 pKa = 4.36KK601 pKa = 9.13TASAATVNAGEE612 pKa = 4.26TLTYY616 pKa = 8.97TLRR619 pKa = 11.84IVNDD623 pKa = 4.38GPATSTSVTLTDD635 pKa = 3.41VLSNLFTNSGGNSFQGVTVNNGTASGGSCGSSGNVGNPSSRR676 pKa = 11.84TLSCTFTSIPVCSGGDD692 pKa = 3.74CPTVTVTILPHH703 pKa = 6.31GNSADD708 pKa = 3.53GTALTRR714 pKa = 11.84GNTASAYY721 pKa = 9.57SSVVADD727 pKa = 3.96PDD729 pKa = 3.81YY730 pKa = 11.78SNNTDD735 pKa = 3.48NVDD738 pKa = 3.36VSIDD742 pKa = 3.58ALTDD746 pKa = 3.58LALSKK751 pKa = 9.68TVTTWSGEE759 pKa = 3.73LGTPLNYY766 pKa = 10.18RR767 pKa = 11.84IQVTNEE773 pKa = 3.73GASGASSLTMTDD785 pKa = 3.56VLPHH789 pKa = 7.17DD790 pKa = 4.07LTYY793 pKa = 11.24LSVNPGGGASCATQPTANSTTEE815 pKa = 3.62ASNDD819 pKa = 3.39QIICTRR825 pKa = 11.84STLARR830 pKa = 11.84GATWTIDD837 pKa = 3.05VATRR841 pKa = 11.84PNHH844 pKa = 6.39GIAPGSTLVNNAVVTTDD861 pKa = 3.45TPEE864 pKa = 4.04TTSDD868 pKa = 3.62NNADD872 pKa = 3.63TADD875 pKa = 3.88AEE877 pKa = 4.36VGEE880 pKa = 4.5ASVDD884 pKa = 3.74LAVIKK889 pKa = 10.59SDD891 pKa = 3.38TPDD894 pKa = 3.38PVFVADD900 pKa = 3.66TVTYY904 pKa = 8.76TVQIINNGPSVASAATIYY922 pKa = 11.0DD923 pKa = 3.99FLPSEE928 pKa = 4.38GFRR931 pKa = 11.84FVEE934 pKa = 5.26DD935 pKa = 3.67SITYY939 pKa = 8.52YY940 pKa = 10.69DD941 pKa = 3.35VSGGVFTEE949 pKa = 4.33IDD951 pKa = 3.69PGDD954 pKa = 3.67LAGLGIACSKK964 pKa = 10.78EE965 pKa = 3.8PDD967 pKa = 3.64DD968 pKa = 4.0EE969 pKa = 5.58AFGTGYY975 pKa = 9.7PDD977 pKa = 3.53QPLDD981 pKa = 5.49DD982 pKa = 4.56SYY984 pKa = 11.36LWPMDD989 pKa = 3.48TVEE992 pKa = 4.78NPDD995 pKa = 3.68YY996 pKa = 11.26LDD998 pKa = 4.5GVWDD1002 pKa = 4.22PLQGDD1007 pKa = 4.09LVEE1010 pKa = 5.7DD1011 pKa = 4.32ADD1013 pKa = 5.34LICNMGLLLNGQIRR1027 pKa = 11.84AIRR1030 pKa = 11.84YY1031 pKa = 7.25EE1032 pKa = 4.05LEE1034 pKa = 4.13ADD1036 pKa = 3.26TRR1038 pKa = 11.84GVYY1041 pKa = 10.26FNYY1044 pKa = 10.44AISRR1048 pKa = 11.84AQEE1051 pKa = 3.87HH1052 pKa = 7.06RR1053 pKa = 11.84DD1054 pKa = 3.54NGADD1058 pKa = 3.73GPDD1061 pKa = 3.58PVPTNDD1067 pKa = 2.87VTRR1070 pKa = 11.84EE1071 pKa = 3.75RR1072 pKa = 11.84TTVRR1076 pKa = 11.84SVPDD1080 pKa = 3.06VSITKK1085 pKa = 9.38VASSSLVSVMEE1096 pKa = 4.3PFDD1099 pKa = 4.11FVITFTNHH1107 pKa = 5.97HH1108 pKa = 5.9ATEE1111 pKa = 3.99PAYY1114 pKa = 10.36FPEE1117 pKa = 5.22IRR1119 pKa = 11.84DD1120 pKa = 3.61VLPAGMEE1127 pKa = 4.05LTGAPILVSGAPGGSVCTGAAGDD1150 pKa = 4.04EE1151 pKa = 4.24SFEE1154 pKa = 4.49CSFGSGLPPGQSVTVSVPVWVVSDD1178 pKa = 3.74GAGPRR1183 pKa = 11.84TNRR1186 pKa = 11.84VEE1188 pKa = 4.06LHH1190 pKa = 6.97LDD1192 pKa = 3.09TDD1194 pKa = 4.41LEE1196 pKa = 4.34FDD1198 pKa = 3.9EE1199 pKa = 5.54PPPPVAEE1206 pKa = 5.13DD1207 pKa = 3.84EE1208 pKa = 4.45DD1209 pKa = 4.6TVDD1212 pKa = 4.07VVISSLAGRR1221 pKa = 11.84VYY1223 pKa = 10.79HH1224 pKa = 7.26DD1225 pKa = 3.55NDD1227 pKa = 3.38YY1228 pKa = 11.56SGVHH1232 pKa = 5.17TVGNPPIEE1240 pKa = 4.27NVLITLNGVSTWGDD1254 pKa = 3.35PVTRR1258 pKa = 11.84SATTAPDD1265 pKa = 3.32GTYY1268 pKa = 10.56FFGDD1272 pKa = 4.34LPSGLYY1278 pKa = 9.26TVTQTQPGGWIDD1290 pKa = 4.08GPVNPGSVGGVPVTNEE1306 pKa = 3.45IGSIEE1311 pKa = 4.28LPADD1315 pKa = 3.48TDD1317 pKa = 3.66AVQYY1321 pKa = 11.4NFGEE1325 pKa = 4.36YY1326 pKa = 10.14LDD1328 pKa = 4.64SDD1330 pKa = 4.19SGQLASVAGHH1340 pKa = 6.69VYY1342 pKa = 10.68HH1343 pKa = 7.55DD1344 pKa = 3.91ANRR1347 pKa = 11.84NGVFDD1352 pKa = 4.55DD1353 pKa = 4.55AEE1355 pKa = 4.33SGIADD1360 pKa = 3.33VVVTLWRR1367 pKa = 11.84DD1368 pKa = 3.32GAVVATALTDD1378 pKa = 3.24AGGAYY1383 pKa = 10.42FFGQLAPGTYY1393 pKa = 9.8RR1394 pKa = 11.84ITEE1397 pKa = 3.91THH1399 pKa = 6.51PDD1401 pKa = 3.04GWIDD1405 pKa = 3.29GAEE1408 pKa = 3.99AVGVGATEE1416 pKa = 4.49PGVSPEE1422 pKa = 3.77TDD1424 pKa = 3.03VFEE1427 pKa = 5.7SIVLQGGDD1435 pKa = 3.04AAQNYY1440 pKa = 10.03DD1441 pKa = 3.27FGEE1444 pKa = 4.31YY1445 pKa = 9.39TSQDD1449 pKa = 2.96IAPIPTLQFKK1459 pKa = 11.06AMLLLILLMGVYY1471 pKa = 10.58AMYY1474 pKa = 10.48RR1475 pKa = 11.84RR1476 pKa = 11.84RR1477 pKa = 11.84EE1478 pKa = 3.91VV1479 pKa = 2.87
MM1 pKa = 7.98IGIEE5 pKa = 4.22LLQGAARR12 pKa = 11.84RR13 pKa = 11.84EE14 pKa = 4.24SGRR17 pKa = 11.84GVNARR22 pKa = 11.84SAARR26 pKa = 11.84RR27 pKa = 11.84LLAVLRR33 pKa = 11.84GLIFVPGLVLAAGTAMAQNVDD54 pKa = 3.57LVLNHH59 pKa = 7.33DD60 pKa = 4.19LTSAATINASDD71 pKa = 3.53TAAFTVTVDD80 pKa = 3.75NSDD83 pKa = 3.5PTHH86 pKa = 6.57TGDD89 pKa = 3.5ANNVVVTYY97 pKa = 9.91QISATDD103 pKa = 3.85TLLSASPSQGSCSLPPDD120 pKa = 3.59ASNTFTCNFGTIPHH134 pKa = 5.84TTNATLDD141 pKa = 3.67VVVRR145 pKa = 11.84ATGTGVGSFPYY156 pKa = 10.73TMTTNAAVATSSTDD170 pKa = 3.35TEE172 pKa = 4.1ATNNDD177 pKa = 3.37EE178 pKa = 4.22EE179 pKa = 4.57RR180 pKa = 11.84TVTVQEE186 pKa = 4.33GTDD189 pKa = 3.29VGIVLNSSPGAIASGGIWTHH209 pKa = 5.33EE210 pKa = 3.84MVVTNHH216 pKa = 5.72GPIGATNIVAHH227 pKa = 7.06LDD229 pKa = 3.73LPAGAILVGSLPAGCSAGAPGVVCQIASLALSGVQNIGPITTQIAATSGSNLSATALVDD288 pKa = 5.08SLDD291 pKa = 3.72QYY293 pKa = 11.88DD294 pKa = 4.51GNTTNQSSTRR304 pKa = 11.84QVDD307 pKa = 3.67VNPGSDD313 pKa = 3.26LAVTLGQSGGPFLEE327 pKa = 4.4NEE329 pKa = 4.25PFALTANPSYY339 pKa = 10.17TGEE342 pKa = 4.2VPDD345 pKa = 4.19VPEE348 pKa = 4.64LSISVDD354 pKa = 3.08PGLQIQNPAVFNQNGWTCSVSGQLVSCSGLDD385 pKa = 3.57TSGMTAGANQPLGVVTVNVVGDD407 pKa = 3.53TAGTYY412 pKa = 9.48SANEE416 pKa = 3.94ATVSADD422 pKa = 3.44YY423 pKa = 10.54TNKK426 pKa = 9.74PDD428 pKa = 4.37PNLDD432 pKa = 3.54NNIDD436 pKa = 3.6GTAPAIVAAEE446 pKa = 4.1LNLGIVKK453 pKa = 10.14SAPQRR458 pKa = 11.84DD459 pKa = 3.91LRR461 pKa = 11.84TIGAAYY467 pKa = 9.18PFDD470 pKa = 3.94YY471 pKa = 10.89SVTVRR476 pKa = 11.84NNGNVGFWGDD486 pKa = 3.93LTWTDD491 pKa = 3.94TVPEE495 pKa = 4.58GLTITNIAAPGGWTCTTVPPNVVGADD521 pKa = 3.22TFTCSRR527 pKa = 11.84TYY529 pKa = 11.43AEE531 pKa = 4.52GAPFAVNATQAFTITAFANGDD552 pKa = 3.41IVGDD556 pKa = 4.08LVNGACLTTIDD567 pKa = 4.43HH568 pKa = 7.51DD569 pKa = 4.86LGFLEE574 pKa = 5.28NEE576 pKa = 4.26VATPCSNATINAQTPGSSADD596 pKa = 3.7LLVEE600 pKa = 4.36KK601 pKa = 9.13TASAATVNAGEE612 pKa = 4.26TLTYY616 pKa = 8.97TLRR619 pKa = 11.84IVNDD623 pKa = 4.38GPATSTSVTLTDD635 pKa = 3.41VLSNLFTNSGGNSFQGVTVNNGTASGGSCGSSGNVGNPSSRR676 pKa = 11.84TLSCTFTSIPVCSGGDD692 pKa = 3.74CPTVTVTILPHH703 pKa = 6.31GNSADD708 pKa = 3.53GTALTRR714 pKa = 11.84GNTASAYY721 pKa = 9.57SSVVADD727 pKa = 3.96PDD729 pKa = 3.81YY730 pKa = 11.78SNNTDD735 pKa = 3.48NVDD738 pKa = 3.36VSIDD742 pKa = 3.58ALTDD746 pKa = 3.58LALSKK751 pKa = 9.68TVTTWSGEE759 pKa = 3.73LGTPLNYY766 pKa = 10.18RR767 pKa = 11.84IQVTNEE773 pKa = 3.73GASGASSLTMTDD785 pKa = 3.56VLPHH789 pKa = 7.17DD790 pKa = 4.07LTYY793 pKa = 11.24LSVNPGGGASCATQPTANSTTEE815 pKa = 3.62ASNDD819 pKa = 3.39QIICTRR825 pKa = 11.84STLARR830 pKa = 11.84GATWTIDD837 pKa = 3.05VATRR841 pKa = 11.84PNHH844 pKa = 6.39GIAPGSTLVNNAVVTTDD861 pKa = 3.45TPEE864 pKa = 4.04TTSDD868 pKa = 3.62NNADD872 pKa = 3.63TADD875 pKa = 3.88AEE877 pKa = 4.36VGEE880 pKa = 4.5ASVDD884 pKa = 3.74LAVIKK889 pKa = 10.59SDD891 pKa = 3.38TPDD894 pKa = 3.38PVFVADD900 pKa = 3.66TVTYY904 pKa = 8.76TVQIINNGPSVASAATIYY922 pKa = 11.0DD923 pKa = 3.99FLPSEE928 pKa = 4.38GFRR931 pKa = 11.84FVEE934 pKa = 5.26DD935 pKa = 3.67SITYY939 pKa = 8.52YY940 pKa = 10.69DD941 pKa = 3.35VSGGVFTEE949 pKa = 4.33IDD951 pKa = 3.69PGDD954 pKa = 3.67LAGLGIACSKK964 pKa = 10.78EE965 pKa = 3.8PDD967 pKa = 3.64DD968 pKa = 4.0EE969 pKa = 5.58AFGTGYY975 pKa = 9.7PDD977 pKa = 3.53QPLDD981 pKa = 5.49DD982 pKa = 4.56SYY984 pKa = 11.36LWPMDD989 pKa = 3.48TVEE992 pKa = 4.78NPDD995 pKa = 3.68YY996 pKa = 11.26LDD998 pKa = 4.5GVWDD1002 pKa = 4.22PLQGDD1007 pKa = 4.09LVEE1010 pKa = 5.7DD1011 pKa = 4.32ADD1013 pKa = 5.34LICNMGLLLNGQIRR1027 pKa = 11.84AIRR1030 pKa = 11.84YY1031 pKa = 7.25EE1032 pKa = 4.05LEE1034 pKa = 4.13ADD1036 pKa = 3.26TRR1038 pKa = 11.84GVYY1041 pKa = 10.26FNYY1044 pKa = 10.44AISRR1048 pKa = 11.84AQEE1051 pKa = 3.87HH1052 pKa = 7.06RR1053 pKa = 11.84DD1054 pKa = 3.54NGADD1058 pKa = 3.73GPDD1061 pKa = 3.58PVPTNDD1067 pKa = 2.87VTRR1070 pKa = 11.84EE1071 pKa = 3.75RR1072 pKa = 11.84TTVRR1076 pKa = 11.84SVPDD1080 pKa = 3.06VSITKK1085 pKa = 9.38VASSSLVSVMEE1096 pKa = 4.3PFDD1099 pKa = 4.11FVITFTNHH1107 pKa = 5.97HH1108 pKa = 5.9ATEE1111 pKa = 3.99PAYY1114 pKa = 10.36FPEE1117 pKa = 5.22IRR1119 pKa = 11.84DD1120 pKa = 3.61VLPAGMEE1127 pKa = 4.05LTGAPILVSGAPGGSVCTGAAGDD1150 pKa = 4.04EE1151 pKa = 4.24SFEE1154 pKa = 4.49CSFGSGLPPGQSVTVSVPVWVVSDD1178 pKa = 3.74GAGPRR1183 pKa = 11.84TNRR1186 pKa = 11.84VEE1188 pKa = 4.06LHH1190 pKa = 6.97LDD1192 pKa = 3.09TDD1194 pKa = 4.41LEE1196 pKa = 4.34FDD1198 pKa = 3.9EE1199 pKa = 5.54PPPPVAEE1206 pKa = 5.13DD1207 pKa = 3.84EE1208 pKa = 4.45DD1209 pKa = 4.6TVDD1212 pKa = 4.07VVISSLAGRR1221 pKa = 11.84VYY1223 pKa = 10.79HH1224 pKa = 7.26DD1225 pKa = 3.55NDD1227 pKa = 3.38YY1228 pKa = 11.56SGVHH1232 pKa = 5.17TVGNPPIEE1240 pKa = 4.27NVLITLNGVSTWGDD1254 pKa = 3.35PVTRR1258 pKa = 11.84SATTAPDD1265 pKa = 3.32GTYY1268 pKa = 10.56FFGDD1272 pKa = 4.34LPSGLYY1278 pKa = 9.26TVTQTQPGGWIDD1290 pKa = 4.08GPVNPGSVGGVPVTNEE1306 pKa = 3.45IGSIEE1311 pKa = 4.28LPADD1315 pKa = 3.48TDD1317 pKa = 3.66AVQYY1321 pKa = 11.4NFGEE1325 pKa = 4.36YY1326 pKa = 10.14LDD1328 pKa = 4.64SDD1330 pKa = 4.19SGQLASVAGHH1340 pKa = 6.69VYY1342 pKa = 10.68HH1343 pKa = 7.55DD1344 pKa = 3.91ANRR1347 pKa = 11.84NGVFDD1352 pKa = 4.55DD1353 pKa = 4.55AEE1355 pKa = 4.33SGIADD1360 pKa = 3.33VVVTLWRR1367 pKa = 11.84DD1368 pKa = 3.32GAVVATALTDD1378 pKa = 3.24AGGAYY1383 pKa = 10.42FFGQLAPGTYY1393 pKa = 9.8RR1394 pKa = 11.84ITEE1397 pKa = 3.91THH1399 pKa = 6.51PDD1401 pKa = 3.04GWIDD1405 pKa = 3.29GAEE1408 pKa = 3.99AVGVGATEE1416 pKa = 4.49PGVSPEE1422 pKa = 3.77TDD1424 pKa = 3.03VFEE1427 pKa = 5.7SIVLQGGDD1435 pKa = 3.04AAQNYY1440 pKa = 10.03DD1441 pKa = 3.27FGEE1444 pKa = 4.31YY1445 pKa = 9.39TSQDD1449 pKa = 2.96IAPIPTLQFKK1459 pKa = 11.06AMLLLILLMGVYY1471 pKa = 10.58AMYY1474 pKa = 10.48RR1475 pKa = 11.84RR1476 pKa = 11.84RR1477 pKa = 11.84EE1478 pKa = 3.91VV1479 pKa = 2.87
Molecular weight: 152.81 kDa
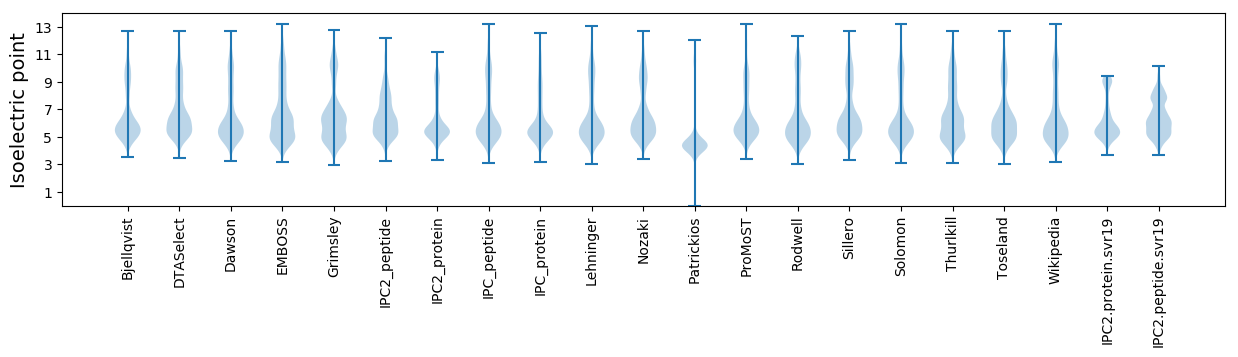
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I6SAJ6|A0A2I6SAJ6_9RHOO Uncharacterized protein OS=Azoarcus pumilus OX=2067960 GN=C0099_00560 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.6RR14 pKa = 11.84THH16 pKa = 5.75GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.91GRR39 pKa = 11.84HH40 pKa = 4.92RR41 pKa = 11.84LAVV44 pKa = 3.37
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.6RR14 pKa = 11.84THH16 pKa = 5.75GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.91GRR39 pKa = 11.84HH40 pKa = 4.92RR41 pKa = 11.84LAVV44 pKa = 3.37
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
971649 |
28 |
1859 |
319.5 |
34.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.797 ± 0.056 | 1.01 ± 0.015 |
5.93 ± 0.035 | 6.245 ± 0.036 |
3.616 ± 0.026 | 8.278 ± 0.041 |
2.41 ± 0.022 | 4.69 ± 0.029 |
2.452 ± 0.032 | 10.672 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.418 ± 0.021 | 2.334 ± 0.023 |
4.976 ± 0.03 | 3.023 ± 0.02 |
8.169 ± 0.049 | 4.895 ± 0.027 |
4.769 ± 0.028 | 7.796 ± 0.035 |
1.393 ± 0.017 | 2.127 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
