
Gloeothece verrucosa (strain PCC 7822) (Cyanothece sp. (strain PCC 7822))
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Chroococcales; Aphanothecaceae; Gloeothece; Gloeothece verrucosa
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
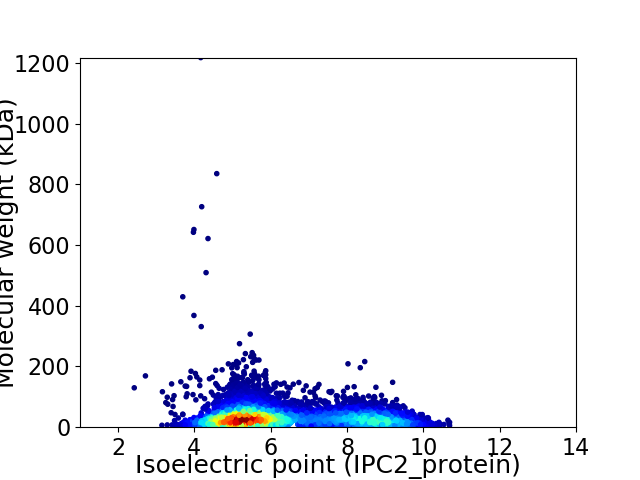
Virtual 2D-PAGE plot for 6559 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E0U6M3|E0U6M3_GLOV7 DEAD/DEAH box helicase domain protein OS=Gloeothece verrucosa (strain PCC 7822) OX=497965 GN=Cyan7822_2822 PE=4 SV=1
MM1 pKa = 7.0ITMNATIYY9 pKa = 10.71RR10 pKa = 11.84NTNADD15 pKa = 3.32SRR17 pKa = 11.84PVSNQLIVKK26 pKa = 7.67LKK28 pKa = 10.64PEE30 pKa = 4.17VQSTQALDD38 pKa = 3.66LQKK41 pKa = 11.24SLGASVIEE49 pKa = 4.43TTKK52 pKa = 10.01QWGYY56 pKa = 8.29QLWSVTGMSIQAALAEE72 pKa = 4.5LKK74 pKa = 10.66KK75 pKa = 10.91NPNVEE80 pKa = 4.13YY81 pKa = 11.27AEE83 pKa = 4.23LNYY86 pKa = 10.76LLEE89 pKa = 4.59TSSIPNDD96 pKa = 3.23VNFNKK101 pKa = 10.14LWAINNTGQTGGTPDD116 pKa = 3.83ADD118 pKa = 3.33IDD120 pKa = 3.83GLEE123 pKa = 4.66AYY125 pKa = 10.21NLANGEE131 pKa = 4.3GVVVGVIDD139 pKa = 3.5TGVDD143 pKa = 3.4YY144 pKa = 8.2THH146 pKa = 7.75PDD148 pKa = 3.18LVNNMWTNPGEE159 pKa = 4.28VPNNGIDD166 pKa = 3.93DD167 pKa = 4.85DD168 pKa = 5.15GNGYY172 pKa = 9.53IDD174 pKa = 5.56DD175 pKa = 4.17YY176 pKa = 11.36YY177 pKa = 11.52GYY179 pKa = 10.99DD180 pKa = 3.74FVNEE184 pKa = 4.37DD185 pKa = 4.65SDD187 pKa = 4.34PLDD190 pKa = 3.72EE191 pKa = 5.22NYY193 pKa = 10.57HH194 pKa = 4.98GTHH197 pKa = 5.35VAGIIAAEE205 pKa = 4.21GNNLQGVIGVAPKK218 pKa = 9.84TKK220 pKa = 10.1IMALKK225 pKa = 9.82FLNGQEE231 pKa = 4.02TGNIFDD237 pKa = 5.84AIQAIEE243 pKa = 4.38YY244 pKa = 8.48ATMMKK249 pKa = 10.36LNYY252 pKa = 9.1GVNVQILNNSWSGGQYY268 pKa = 10.15SQALHH273 pKa = 6.69DD274 pKa = 5.79AIAAANAAGQLFTASAGNGHH294 pKa = 6.5NNNDD298 pKa = 4.22LLPVYY303 pKa = 9.13PASYY307 pKa = 10.37DD308 pKa = 3.47LEE310 pKa = 4.75NIISVAATDD319 pKa = 5.12DD320 pKa = 3.65QDD322 pKa = 3.86QLADD326 pKa = 4.2FSNYY330 pKa = 9.67GATTVDD336 pKa = 3.88LAAPGVQIYY345 pKa = 7.48STWPGNNYY353 pKa = 10.01ASSSGTSMSTAYY365 pKa = 10.55VSGVASLIYY374 pKa = 10.42SLNPNLTPAQVKK386 pKa = 10.19SYY388 pKa = 9.66ILSGVEE394 pKa = 4.23PVSSLQGITVSGGRR408 pKa = 11.84LNAYY412 pKa = 8.66KK413 pKa = 10.78ALLSPLAAKK422 pKa = 8.69ITGSLWNDD430 pKa = 3.32LDD432 pKa = 3.99QNGVRR437 pKa = 11.84DD438 pKa = 3.72AGEE441 pKa = 4.34PGLAGWKK448 pKa = 9.81IYY450 pKa = 10.79LDD452 pKa = 3.59QNNNGTFDD460 pKa = 3.7PGEE463 pKa = 4.13RR464 pKa = 11.84STVTNKK470 pKa = 10.19QGDD473 pKa = 3.95YY474 pKa = 11.33AFLLLSPGNYY484 pKa = 7.08TVRR487 pKa = 11.84EE488 pKa = 4.15VKK490 pKa = 10.36QLGWTQTNQTDD501 pKa = 4.01LYY503 pKa = 8.89YY504 pKa = 9.48TVQVAEE510 pKa = 4.31SQTVSGINWGNFLAQPATISGTKK533 pKa = 9.54WNDD536 pKa = 3.03LNQNAVLDD544 pKa = 3.97SGEE547 pKa = 4.43PGLAGWTIYY556 pKa = 10.84LDD558 pKa = 3.78EE559 pKa = 5.2NGNSQLDD566 pKa = 3.65TGEE569 pKa = 4.53LSTVTDD575 pKa = 3.63EE576 pKa = 3.86QGKK579 pKa = 10.38YY580 pKa = 10.68SFTNLAPGNYY590 pKa = 8.41QVNVVNQNGWQQTQPVSGSYY610 pKa = 10.35SVALSPDD617 pKa = 3.11EE618 pKa = 3.99VVNNINFGHH627 pKa = 6.04YY628 pKa = 7.77EE629 pKa = 3.99LKK631 pKa = 10.49KK632 pKa = 10.98GEE634 pKa = 3.75IRR636 pKa = 11.84GIKK639 pKa = 9.3WNDD642 pKa = 3.16LDD644 pKa = 4.6GDD646 pKa = 4.48GIRR649 pKa = 11.84DD650 pKa = 3.46QGEE653 pKa = 3.95PGLPGMTIYY662 pKa = 11.02LDD664 pKa = 3.47QNQNGRR670 pKa = 11.84LDD672 pKa = 3.6NGEE675 pKa = 3.9ISALTDD681 pKa = 3.12KK682 pKa = 11.04DD683 pKa = 3.41GNYY686 pKa = 10.8AFTNLTPGITYY697 pKa = 8.57TVAEE701 pKa = 4.25VLQANWAQTTPRR713 pKa = 11.84FLFNADD719 pKa = 3.18FSDD722 pKa = 3.86AQGNASLDD730 pKa = 3.37GFTIDD735 pKa = 4.58NNYY738 pKa = 9.86NGLWHH743 pKa = 7.22LSTGRR748 pKa = 11.84GNQAGHH754 pKa = 6.19SADD757 pKa = 3.46DD758 pKa = 3.5SLYY761 pKa = 10.25FGTKK765 pKa = 9.69EE766 pKa = 4.35GIDD769 pKa = 3.29GGGNYY774 pKa = 10.19DD775 pKa = 3.42VGSTSGRR782 pKa = 11.84ITSSEE787 pKa = 3.4ISLIGLEE794 pKa = 4.18DD795 pKa = 3.58AQLSFNYY802 pKa = 9.8FLQTEE807 pKa = 4.24GGGFYY812 pKa = 10.83DD813 pKa = 3.38QAKK816 pKa = 10.37VLLSQNGGDD825 pKa = 3.97FFQIASNQQSLQDD838 pKa = 4.06PTPGWTNLTLDD849 pKa = 4.0LTAYY853 pKa = 8.37TGSKK857 pKa = 9.6IKK859 pKa = 10.31IRR861 pKa = 11.84FDD863 pKa = 3.54FDD865 pKa = 4.73SIDD868 pKa = 3.54YY869 pKa = 9.41YY870 pKa = 11.89ANNFEE875 pKa = 4.08GWYY878 pKa = 9.64IDD880 pKa = 5.73DD881 pKa = 3.58ITVTNAAGQGIYY893 pKa = 7.63TVKK896 pKa = 10.47LDD898 pKa = 3.77PGQIVNNINFGNYY911 pKa = 7.61QPVKK915 pKa = 10.41NGTWRR920 pKa = 11.84NDD922 pKa = 3.22EE923 pKa = 4.92LIGTKK928 pKa = 10.35LKK930 pKa = 10.83DD931 pKa = 3.86IINGLAGNDD940 pKa = 3.34NLYY943 pKa = 10.97GKK945 pKa = 9.97EE946 pKa = 4.33DD947 pKa = 3.85NDD949 pKa = 3.66VLNGGGGNDD958 pKa = 3.04NLYY961 pKa = 11.1GNEE964 pKa = 4.59GNDD967 pKa = 3.28EE968 pKa = 4.41LNGNEE973 pKa = 4.06GKK975 pKa = 10.26DD976 pKa = 3.67YY977 pKa = 11.73LEE979 pKa = 4.73GNQGNDD985 pKa = 3.09SLNGGSEE992 pKa = 4.05SDD994 pKa = 3.36NLLGNEE1000 pKa = 4.79GNDD1003 pKa = 3.54TLKK1006 pKa = 11.14GGTHH1010 pKa = 7.18NDD1012 pKa = 4.06TLWGNQGNDD1021 pKa = 3.15ILNGDD1026 pKa = 4.19EE1027 pKa = 5.68GNDD1030 pKa = 3.46LLYY1033 pKa = 11.16GNEE1036 pKa = 4.22GQDD1039 pKa = 3.53TLKK1042 pKa = 11.05GGSNNDD1048 pKa = 3.28TLYY1051 pKa = 11.37GNEE1054 pKa = 4.66DD1055 pKa = 3.53NDD1057 pKa = 4.29LLSGDD1062 pKa = 4.01EE1063 pKa = 5.38SNDD1066 pKa = 3.28QLYY1069 pKa = 10.87GDD1071 pKa = 4.3QGNDD1075 pKa = 3.11ILNGDD1080 pKa = 4.3AGNDD1084 pKa = 3.38ALYY1087 pKa = 10.82GSTGNDD1093 pKa = 3.24TLSGGVGRR1101 pKa = 11.84DD1102 pKa = 3.17TLYY1105 pKa = 11.43GNEE1108 pKa = 4.53NNDD1111 pKa = 4.55LLNGDD1116 pKa = 4.88DD1117 pKa = 5.65DD1118 pKa = 5.76HH1119 pKa = 7.97DD1120 pKa = 4.3LLYY1123 pKa = 11.21GNQDD1127 pKa = 2.96NDD1129 pKa = 3.87TLNGGNGNDD1138 pKa = 3.27TLYY1141 pKa = 11.19GNEE1144 pKa = 4.67GSDD1147 pKa = 4.2ILNGGNNNDD1156 pKa = 3.34SLYY1159 pKa = 11.11GNEE1162 pKa = 5.46GNDD1165 pKa = 3.68TLNGAEE1171 pKa = 4.58GNDD1174 pKa = 3.62TLRR1177 pKa = 11.84GGEE1180 pKa = 4.3GNDD1183 pKa = 3.86LLTGGLDD1190 pKa = 3.08KK1191 pKa = 11.34DD1192 pKa = 4.35VFVLAANFGSDD1203 pKa = 3.66TITDD1207 pKa = 4.05FMDD1210 pKa = 4.25GQDD1213 pKa = 4.22KK1214 pKa = 11.18LGLSAGLSFAQLTLTSNQSNTLIYY1238 pKa = 9.26LTSSNEE1244 pKa = 3.88LLATLSGINLSLINSRR1260 pKa = 11.84DD1261 pKa = 3.4FMVII1265 pKa = 3.31
MM1 pKa = 7.0ITMNATIYY9 pKa = 10.71RR10 pKa = 11.84NTNADD15 pKa = 3.32SRR17 pKa = 11.84PVSNQLIVKK26 pKa = 7.67LKK28 pKa = 10.64PEE30 pKa = 4.17VQSTQALDD38 pKa = 3.66LQKK41 pKa = 11.24SLGASVIEE49 pKa = 4.43TTKK52 pKa = 10.01QWGYY56 pKa = 8.29QLWSVTGMSIQAALAEE72 pKa = 4.5LKK74 pKa = 10.66KK75 pKa = 10.91NPNVEE80 pKa = 4.13YY81 pKa = 11.27AEE83 pKa = 4.23LNYY86 pKa = 10.76LLEE89 pKa = 4.59TSSIPNDD96 pKa = 3.23VNFNKK101 pKa = 10.14LWAINNTGQTGGTPDD116 pKa = 3.83ADD118 pKa = 3.33IDD120 pKa = 3.83GLEE123 pKa = 4.66AYY125 pKa = 10.21NLANGEE131 pKa = 4.3GVVVGVIDD139 pKa = 3.5TGVDD143 pKa = 3.4YY144 pKa = 8.2THH146 pKa = 7.75PDD148 pKa = 3.18LVNNMWTNPGEE159 pKa = 4.28VPNNGIDD166 pKa = 3.93DD167 pKa = 4.85DD168 pKa = 5.15GNGYY172 pKa = 9.53IDD174 pKa = 5.56DD175 pKa = 4.17YY176 pKa = 11.36YY177 pKa = 11.52GYY179 pKa = 10.99DD180 pKa = 3.74FVNEE184 pKa = 4.37DD185 pKa = 4.65SDD187 pKa = 4.34PLDD190 pKa = 3.72EE191 pKa = 5.22NYY193 pKa = 10.57HH194 pKa = 4.98GTHH197 pKa = 5.35VAGIIAAEE205 pKa = 4.21GNNLQGVIGVAPKK218 pKa = 9.84TKK220 pKa = 10.1IMALKK225 pKa = 9.82FLNGQEE231 pKa = 4.02TGNIFDD237 pKa = 5.84AIQAIEE243 pKa = 4.38YY244 pKa = 8.48ATMMKK249 pKa = 10.36LNYY252 pKa = 9.1GVNVQILNNSWSGGQYY268 pKa = 10.15SQALHH273 pKa = 6.69DD274 pKa = 5.79AIAAANAAGQLFTASAGNGHH294 pKa = 6.5NNNDD298 pKa = 4.22LLPVYY303 pKa = 9.13PASYY307 pKa = 10.37DD308 pKa = 3.47LEE310 pKa = 4.75NIISVAATDD319 pKa = 5.12DD320 pKa = 3.65QDD322 pKa = 3.86QLADD326 pKa = 4.2FSNYY330 pKa = 9.67GATTVDD336 pKa = 3.88LAAPGVQIYY345 pKa = 7.48STWPGNNYY353 pKa = 10.01ASSSGTSMSTAYY365 pKa = 10.55VSGVASLIYY374 pKa = 10.42SLNPNLTPAQVKK386 pKa = 10.19SYY388 pKa = 9.66ILSGVEE394 pKa = 4.23PVSSLQGITVSGGRR408 pKa = 11.84LNAYY412 pKa = 8.66KK413 pKa = 10.78ALLSPLAAKK422 pKa = 8.69ITGSLWNDD430 pKa = 3.32LDD432 pKa = 3.99QNGVRR437 pKa = 11.84DD438 pKa = 3.72AGEE441 pKa = 4.34PGLAGWKK448 pKa = 9.81IYY450 pKa = 10.79LDD452 pKa = 3.59QNNNGTFDD460 pKa = 3.7PGEE463 pKa = 4.13RR464 pKa = 11.84STVTNKK470 pKa = 10.19QGDD473 pKa = 3.95YY474 pKa = 11.33AFLLLSPGNYY484 pKa = 7.08TVRR487 pKa = 11.84EE488 pKa = 4.15VKK490 pKa = 10.36QLGWTQTNQTDD501 pKa = 4.01LYY503 pKa = 8.89YY504 pKa = 9.48TVQVAEE510 pKa = 4.31SQTVSGINWGNFLAQPATISGTKK533 pKa = 9.54WNDD536 pKa = 3.03LNQNAVLDD544 pKa = 3.97SGEE547 pKa = 4.43PGLAGWTIYY556 pKa = 10.84LDD558 pKa = 3.78EE559 pKa = 5.2NGNSQLDD566 pKa = 3.65TGEE569 pKa = 4.53LSTVTDD575 pKa = 3.63EE576 pKa = 3.86QGKK579 pKa = 10.38YY580 pKa = 10.68SFTNLAPGNYY590 pKa = 8.41QVNVVNQNGWQQTQPVSGSYY610 pKa = 10.35SVALSPDD617 pKa = 3.11EE618 pKa = 3.99VVNNINFGHH627 pKa = 6.04YY628 pKa = 7.77EE629 pKa = 3.99LKK631 pKa = 10.49KK632 pKa = 10.98GEE634 pKa = 3.75IRR636 pKa = 11.84GIKK639 pKa = 9.3WNDD642 pKa = 3.16LDD644 pKa = 4.6GDD646 pKa = 4.48GIRR649 pKa = 11.84DD650 pKa = 3.46QGEE653 pKa = 3.95PGLPGMTIYY662 pKa = 11.02LDD664 pKa = 3.47QNQNGRR670 pKa = 11.84LDD672 pKa = 3.6NGEE675 pKa = 3.9ISALTDD681 pKa = 3.12KK682 pKa = 11.04DD683 pKa = 3.41GNYY686 pKa = 10.8AFTNLTPGITYY697 pKa = 8.57TVAEE701 pKa = 4.25VLQANWAQTTPRR713 pKa = 11.84FLFNADD719 pKa = 3.18FSDD722 pKa = 3.86AQGNASLDD730 pKa = 3.37GFTIDD735 pKa = 4.58NNYY738 pKa = 9.86NGLWHH743 pKa = 7.22LSTGRR748 pKa = 11.84GNQAGHH754 pKa = 6.19SADD757 pKa = 3.46DD758 pKa = 3.5SLYY761 pKa = 10.25FGTKK765 pKa = 9.69EE766 pKa = 4.35GIDD769 pKa = 3.29GGGNYY774 pKa = 10.19DD775 pKa = 3.42VGSTSGRR782 pKa = 11.84ITSSEE787 pKa = 3.4ISLIGLEE794 pKa = 4.18DD795 pKa = 3.58AQLSFNYY802 pKa = 9.8FLQTEE807 pKa = 4.24GGGFYY812 pKa = 10.83DD813 pKa = 3.38QAKK816 pKa = 10.37VLLSQNGGDD825 pKa = 3.97FFQIASNQQSLQDD838 pKa = 4.06PTPGWTNLTLDD849 pKa = 4.0LTAYY853 pKa = 8.37TGSKK857 pKa = 9.6IKK859 pKa = 10.31IRR861 pKa = 11.84FDD863 pKa = 3.54FDD865 pKa = 4.73SIDD868 pKa = 3.54YY869 pKa = 9.41YY870 pKa = 11.89ANNFEE875 pKa = 4.08GWYY878 pKa = 9.64IDD880 pKa = 5.73DD881 pKa = 3.58ITVTNAAGQGIYY893 pKa = 7.63TVKK896 pKa = 10.47LDD898 pKa = 3.77PGQIVNNINFGNYY911 pKa = 7.61QPVKK915 pKa = 10.41NGTWRR920 pKa = 11.84NDD922 pKa = 3.22EE923 pKa = 4.92LIGTKK928 pKa = 10.35LKK930 pKa = 10.83DD931 pKa = 3.86IINGLAGNDD940 pKa = 3.34NLYY943 pKa = 10.97GKK945 pKa = 9.97EE946 pKa = 4.33DD947 pKa = 3.85NDD949 pKa = 3.66VLNGGGGNDD958 pKa = 3.04NLYY961 pKa = 11.1GNEE964 pKa = 4.59GNDD967 pKa = 3.28EE968 pKa = 4.41LNGNEE973 pKa = 4.06GKK975 pKa = 10.26DD976 pKa = 3.67YY977 pKa = 11.73LEE979 pKa = 4.73GNQGNDD985 pKa = 3.09SLNGGSEE992 pKa = 4.05SDD994 pKa = 3.36NLLGNEE1000 pKa = 4.79GNDD1003 pKa = 3.54TLKK1006 pKa = 11.14GGTHH1010 pKa = 7.18NDD1012 pKa = 4.06TLWGNQGNDD1021 pKa = 3.15ILNGDD1026 pKa = 4.19EE1027 pKa = 5.68GNDD1030 pKa = 3.46LLYY1033 pKa = 11.16GNEE1036 pKa = 4.22GQDD1039 pKa = 3.53TLKK1042 pKa = 11.05GGSNNDD1048 pKa = 3.28TLYY1051 pKa = 11.37GNEE1054 pKa = 4.66DD1055 pKa = 3.53NDD1057 pKa = 4.29LLSGDD1062 pKa = 4.01EE1063 pKa = 5.38SNDD1066 pKa = 3.28QLYY1069 pKa = 10.87GDD1071 pKa = 4.3QGNDD1075 pKa = 3.11ILNGDD1080 pKa = 4.3AGNDD1084 pKa = 3.38ALYY1087 pKa = 10.82GSTGNDD1093 pKa = 3.24TLSGGVGRR1101 pKa = 11.84DD1102 pKa = 3.17TLYY1105 pKa = 11.43GNEE1108 pKa = 4.53NNDD1111 pKa = 4.55LLNGDD1116 pKa = 4.88DD1117 pKa = 5.65DD1118 pKa = 5.76HH1119 pKa = 7.97DD1120 pKa = 4.3LLYY1123 pKa = 11.21GNQDD1127 pKa = 2.96NDD1129 pKa = 3.87TLNGGNGNDD1138 pKa = 3.27TLYY1141 pKa = 11.19GNEE1144 pKa = 4.67GSDD1147 pKa = 4.2ILNGGNNNDD1156 pKa = 3.34SLYY1159 pKa = 11.11GNEE1162 pKa = 5.46GNDD1165 pKa = 3.68TLNGAEE1171 pKa = 4.58GNDD1174 pKa = 3.62TLRR1177 pKa = 11.84GGEE1180 pKa = 4.3GNDD1183 pKa = 3.86LLTGGLDD1190 pKa = 3.08KK1191 pKa = 11.34DD1192 pKa = 4.35VFVLAANFGSDD1203 pKa = 3.66TITDD1207 pKa = 4.05FMDD1210 pKa = 4.25GQDD1213 pKa = 4.22KK1214 pKa = 11.18LGLSAGLSFAQLTLTSNQSNTLIYY1238 pKa = 9.26LTSSNEE1244 pKa = 3.88LLATLSGINLSLINSRR1260 pKa = 11.84DD1261 pKa = 3.4FMVII1265 pKa = 3.31
Molecular weight: 135.45 kDa
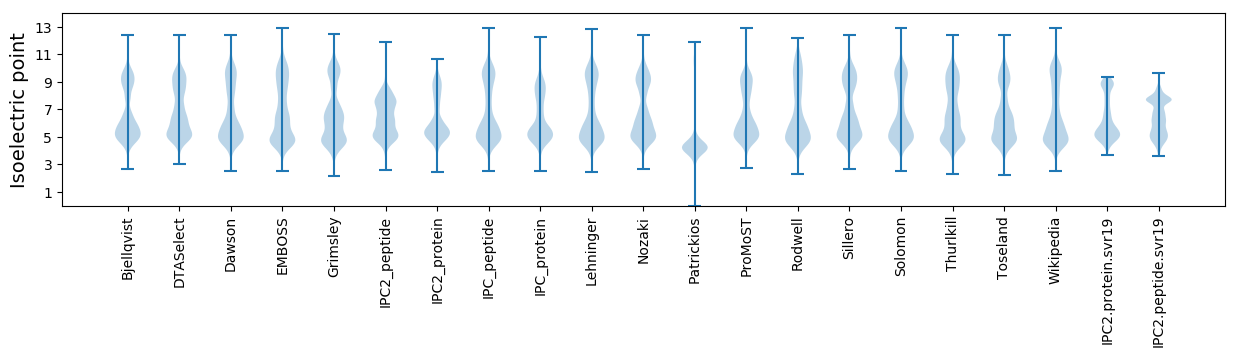
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E0U8M4|E0U8M4_GLOV7 Serine/threonine protein kinase OS=Gloeothece verrucosa (strain PCC 7822) OX=497965 GN=Cyan7822_1783 PE=4 SV=1
MM1 pKa = 7.74RR2 pKa = 11.84RR3 pKa = 11.84LLCYY7 pKa = 10.09QEE9 pKa = 5.72LVFHH13 pKa = 5.43QQHH16 pKa = 6.48YY17 pKa = 10.81IDD19 pKa = 5.25LLNSGQVDD27 pKa = 4.11PLAHH31 pKa = 6.68SLIRR35 pKa = 11.84WHH37 pKa = 6.0IRR39 pKa = 11.84KK40 pKa = 9.48LGRR43 pKa = 11.84EE44 pKa = 3.85IDD46 pKa = 3.98TRR48 pKa = 11.84WPPRR52 pKa = 11.84LRR54 pKa = 11.84KK55 pKa = 9.64VHH57 pKa = 5.09VV58 pKa = 3.53
MM1 pKa = 7.74RR2 pKa = 11.84RR3 pKa = 11.84LLCYY7 pKa = 10.09QEE9 pKa = 5.72LVFHH13 pKa = 5.43QQHH16 pKa = 6.48YY17 pKa = 10.81IDD19 pKa = 5.25LLNSGQVDD27 pKa = 4.11PLAHH31 pKa = 6.68SLIRR35 pKa = 11.84WHH37 pKa = 6.0IRR39 pKa = 11.84KK40 pKa = 9.48LGRR43 pKa = 11.84EE44 pKa = 3.85IDD46 pKa = 3.98TRR48 pKa = 11.84WPPRR52 pKa = 11.84LRR54 pKa = 11.84KK55 pKa = 9.64VHH57 pKa = 5.09VV58 pKa = 3.53
Molecular weight: 7.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2140918 |
30 |
11342 |
326.4 |
36.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.295 ± 0.033 | 0.98 ± 0.012 |
4.826 ± 0.033 | 6.522 ± 0.042 |
4.062 ± 0.023 | 6.458 ± 0.041 |
1.758 ± 0.018 | 7.158 ± 0.026 |
5.504 ± 0.037 | 11.158 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.703 ± 0.018 | 4.829 ± 0.043 |
4.716 ± 0.025 | 5.404 ± 0.032 |
4.608 ± 0.033 | 6.549 ± 0.03 |
5.641 ± 0.049 | 6.036 ± 0.026 |
1.459 ± 0.015 | 3.333 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
