
Alces alces faeces associated microvirus MP12 5423
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
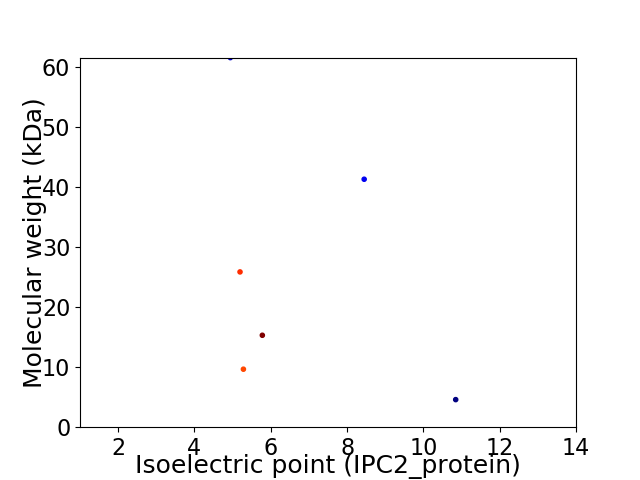
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CL36|A0A2Z5CL36_9VIRU Minor capsid protein OS=Alces alces faeces associated microvirus MP12 5423 OX=2219135 PE=4 SV=1
MM1 pKa = 7.36NKK3 pKa = 9.59YY4 pKa = 9.73FATNPRR10 pKa = 11.84IKK12 pKa = 10.05ISRR15 pKa = 11.84TKK17 pKa = 10.25FDD19 pKa = 4.03LSSTHH24 pKa = 5.8KK25 pKa = 7.65TTFNAGYY32 pKa = 9.53LVPIKK37 pKa = 10.2IYY39 pKa = 10.77EE40 pKa = 4.17NVPGDD45 pKa = 4.02TISVSMNSVIEE56 pKa = 4.0MTTPLKK62 pKa = 9.78PTMDD66 pKa = 3.75LAVCNVYY73 pKa = 10.6AFKK76 pKa = 11.33VPMRR80 pKa = 11.84LLWEE84 pKa = 4.31HH85 pKa = 4.69WQEE88 pKa = 4.05MFGEE92 pKa = 4.21NDD94 pKa = 3.2DD95 pKa = 4.63TYY97 pKa = 9.75WAQPVEE103 pKa = 4.08YY104 pKa = 9.03TIPKK108 pKa = 8.31LTAPTGGWSKK118 pKa = 11.3DD119 pKa = 3.35SIMSHH124 pKa = 5.59MGVRR128 pKa = 11.84MNTEE132 pKa = 4.06GLSVSALPARR142 pKa = 11.84AVAKK146 pKa = 10.17IYY148 pKa = 10.91NDD150 pKa = 3.09WFRR153 pKa = 11.84DD154 pKa = 3.77QNVMTPYY161 pKa = 10.93LEE163 pKa = 5.38AKK165 pKa = 10.08DD166 pKa = 3.68DD167 pKa = 4.24SNRR170 pKa = 11.84EE171 pKa = 3.77GSNGDD176 pKa = 3.38GKK178 pKa = 10.76NDD180 pKa = 3.3LVAGGKK186 pKa = 9.39CPRR189 pKa = 11.84IAKK192 pKa = 9.63FHH194 pKa = 7.28DD195 pKa = 3.69IFTTALPGPIKK206 pKa = 10.16STEE209 pKa = 4.05DD210 pKa = 3.11VLLPLGEE217 pKa = 4.17FAPVGTRR224 pKa = 11.84TNVSPQLTNNTMLTWRR240 pKa = 11.84MSNGEE245 pKa = 4.26SLTSGTSYY253 pKa = 11.14NITIGANDD261 pKa = 3.79YY262 pKa = 10.07TPTQARR268 pKa = 11.84TDD270 pKa = 3.6VDD272 pKa = 4.26LGTYY276 pKa = 9.95SADD279 pKa = 3.28IQPKK283 pKa = 9.63NLWADD288 pKa = 3.92LSQATGATMQDD299 pKa = 3.07FLYY302 pKa = 10.42AYY304 pKa = 9.86SLYY307 pKa = 11.42KK308 pKa = 10.11MFTIDD313 pKa = 3.02ARR315 pKa = 11.84GGTRR319 pKa = 11.84YY320 pKa = 8.58TEE322 pKa = 3.81ILANHH327 pKa = 6.39YY328 pKa = 10.64GVISSDD334 pKa = 3.0ARR336 pKa = 11.84LQRR339 pKa = 11.84SEE341 pKa = 4.16YY342 pKa = 10.8LGGTNFEE349 pKa = 4.51INMVHH354 pKa = 6.39VPQTSQTTEE363 pKa = 3.78EE364 pKa = 4.36SAQGYY369 pKa = 7.15LTAYY373 pKa = 10.4SNTAIRR379 pKa = 11.84NKK381 pKa = 10.48HH382 pKa = 6.06LFTTSYY388 pKa = 11.24DD389 pKa = 3.57EE390 pKa = 4.02HH391 pKa = 8.22CYY393 pKa = 10.27TILFAAIRR401 pKa = 11.84TKK403 pKa = 8.55QTYY406 pKa = 9.1AQGIEE411 pKa = 4.06KK412 pKa = 8.89MWFHH416 pKa = 6.88EE417 pKa = 4.18EE418 pKa = 3.78RR419 pKa = 11.84TDD421 pKa = 3.89FYY423 pKa = 10.58MPTFAHH429 pKa = 6.88IGEE432 pKa = 4.1QPIRR436 pKa = 11.84NRR438 pKa = 11.84EE439 pKa = 3.97LFAQGTDD446 pKa = 2.93EE447 pKa = 4.64DD448 pKa = 4.17AQTFGFGEE456 pKa = 3.92AFYY459 pKa = 10.28EE460 pKa = 4.16YY461 pKa = 8.97RR462 pKa = 11.84TQVDD466 pKa = 4.39LVTGEE471 pKa = 4.41FSPDD475 pKa = 3.28AEE477 pKa = 4.3NSLPSYY483 pKa = 10.39TYY485 pKa = 10.2TNDD488 pKa = 3.4FEE490 pKa = 5.15SLPTLSEE497 pKa = 3.95EE498 pKa = 4.93FILEE502 pKa = 4.13TEE504 pKa = 4.41DD505 pKa = 3.5NVNRR509 pKa = 11.84TIAVNSSLQDD519 pKa = 3.1QFKK522 pKa = 11.08ADD524 pKa = 4.07FYY526 pKa = 11.43FNVTAVRR533 pKa = 11.84PLPPRR538 pKa = 11.84SIPSLASHH546 pKa = 7.25FF547 pKa = 4.11
MM1 pKa = 7.36NKK3 pKa = 9.59YY4 pKa = 9.73FATNPRR10 pKa = 11.84IKK12 pKa = 10.05ISRR15 pKa = 11.84TKK17 pKa = 10.25FDD19 pKa = 4.03LSSTHH24 pKa = 5.8KK25 pKa = 7.65TTFNAGYY32 pKa = 9.53LVPIKK37 pKa = 10.2IYY39 pKa = 10.77EE40 pKa = 4.17NVPGDD45 pKa = 4.02TISVSMNSVIEE56 pKa = 4.0MTTPLKK62 pKa = 9.78PTMDD66 pKa = 3.75LAVCNVYY73 pKa = 10.6AFKK76 pKa = 11.33VPMRR80 pKa = 11.84LLWEE84 pKa = 4.31HH85 pKa = 4.69WQEE88 pKa = 4.05MFGEE92 pKa = 4.21NDD94 pKa = 3.2DD95 pKa = 4.63TYY97 pKa = 9.75WAQPVEE103 pKa = 4.08YY104 pKa = 9.03TIPKK108 pKa = 8.31LTAPTGGWSKK118 pKa = 11.3DD119 pKa = 3.35SIMSHH124 pKa = 5.59MGVRR128 pKa = 11.84MNTEE132 pKa = 4.06GLSVSALPARR142 pKa = 11.84AVAKK146 pKa = 10.17IYY148 pKa = 10.91NDD150 pKa = 3.09WFRR153 pKa = 11.84DD154 pKa = 3.77QNVMTPYY161 pKa = 10.93LEE163 pKa = 5.38AKK165 pKa = 10.08DD166 pKa = 3.68DD167 pKa = 4.24SNRR170 pKa = 11.84EE171 pKa = 3.77GSNGDD176 pKa = 3.38GKK178 pKa = 10.76NDD180 pKa = 3.3LVAGGKK186 pKa = 9.39CPRR189 pKa = 11.84IAKK192 pKa = 9.63FHH194 pKa = 7.28DD195 pKa = 3.69IFTTALPGPIKK206 pKa = 10.16STEE209 pKa = 4.05DD210 pKa = 3.11VLLPLGEE217 pKa = 4.17FAPVGTRR224 pKa = 11.84TNVSPQLTNNTMLTWRR240 pKa = 11.84MSNGEE245 pKa = 4.26SLTSGTSYY253 pKa = 11.14NITIGANDD261 pKa = 3.79YY262 pKa = 10.07TPTQARR268 pKa = 11.84TDD270 pKa = 3.6VDD272 pKa = 4.26LGTYY276 pKa = 9.95SADD279 pKa = 3.28IQPKK283 pKa = 9.63NLWADD288 pKa = 3.92LSQATGATMQDD299 pKa = 3.07FLYY302 pKa = 10.42AYY304 pKa = 9.86SLYY307 pKa = 11.42KK308 pKa = 10.11MFTIDD313 pKa = 3.02ARR315 pKa = 11.84GGTRR319 pKa = 11.84YY320 pKa = 8.58TEE322 pKa = 3.81ILANHH327 pKa = 6.39YY328 pKa = 10.64GVISSDD334 pKa = 3.0ARR336 pKa = 11.84LQRR339 pKa = 11.84SEE341 pKa = 4.16YY342 pKa = 10.8LGGTNFEE349 pKa = 4.51INMVHH354 pKa = 6.39VPQTSQTTEE363 pKa = 3.78EE364 pKa = 4.36SAQGYY369 pKa = 7.15LTAYY373 pKa = 10.4SNTAIRR379 pKa = 11.84NKK381 pKa = 10.48HH382 pKa = 6.06LFTTSYY388 pKa = 11.24DD389 pKa = 3.57EE390 pKa = 4.02HH391 pKa = 8.22CYY393 pKa = 10.27TILFAAIRR401 pKa = 11.84TKK403 pKa = 8.55QTYY406 pKa = 9.1AQGIEE411 pKa = 4.06KK412 pKa = 8.89MWFHH416 pKa = 6.88EE417 pKa = 4.18EE418 pKa = 3.78RR419 pKa = 11.84TDD421 pKa = 3.89FYY423 pKa = 10.58MPTFAHH429 pKa = 6.88IGEE432 pKa = 4.1QPIRR436 pKa = 11.84NRR438 pKa = 11.84EE439 pKa = 3.97LFAQGTDD446 pKa = 2.93EE447 pKa = 4.64DD448 pKa = 4.17AQTFGFGEE456 pKa = 3.92AFYY459 pKa = 10.28EE460 pKa = 4.16YY461 pKa = 8.97RR462 pKa = 11.84TQVDD466 pKa = 4.39LVTGEE471 pKa = 4.41FSPDD475 pKa = 3.28AEE477 pKa = 4.3NSLPSYY483 pKa = 10.39TYY485 pKa = 10.2TNDD488 pKa = 3.4FEE490 pKa = 5.15SLPTLSEE497 pKa = 3.95EE498 pKa = 4.93FILEE502 pKa = 4.13TEE504 pKa = 4.41DD505 pKa = 3.5NVNRR509 pKa = 11.84TIAVNSSLQDD519 pKa = 3.1QFKK522 pKa = 11.08ADD524 pKa = 4.07FYY526 pKa = 11.43FNVTAVRR533 pKa = 11.84PLPPRR538 pKa = 11.84SIPSLASHH546 pKa = 7.25FF547 pKa = 4.11
Molecular weight: 61.63 kDa
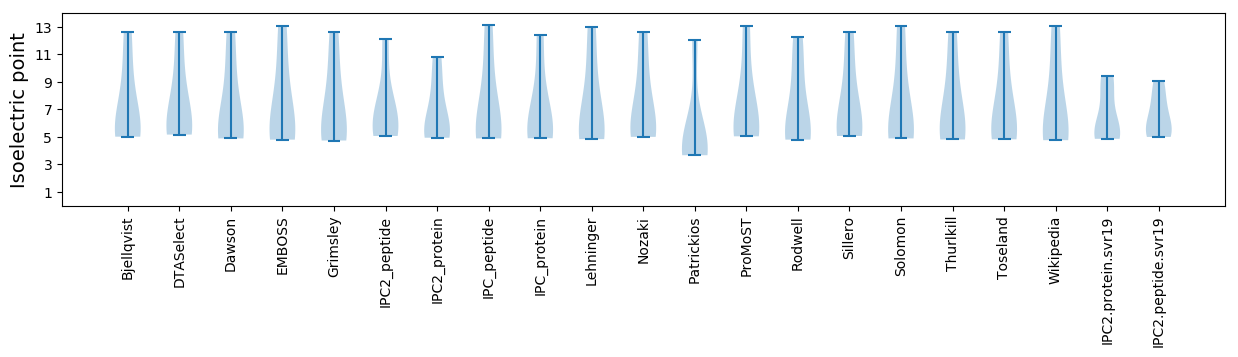
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CJ08|A0A2Z5CJ08_9VIRU Nonstructural protein OS=Alces alces faeces associated microvirus MP12 5423 OX=2219135 PE=4 SV=1
MM1 pKa = 7.55AKK3 pKa = 10.33KK4 pKa = 9.86IRR6 pKa = 11.84NKK8 pKa = 10.41ARR10 pKa = 11.84DD11 pKa = 3.29RR12 pKa = 11.84RR13 pKa = 11.84IFTRR17 pKa = 11.84TASRR21 pKa = 11.84MHH23 pKa = 6.68KK24 pKa = 10.49ANRR27 pKa = 11.84TPRR30 pKa = 11.84FMRR33 pKa = 11.84GGIRR37 pKa = 11.84LL38 pKa = 3.51
MM1 pKa = 7.55AKK3 pKa = 10.33KK4 pKa = 9.86IRR6 pKa = 11.84NKK8 pKa = 10.41ARR10 pKa = 11.84DD11 pKa = 3.29RR12 pKa = 11.84RR13 pKa = 11.84IFTRR17 pKa = 11.84TASRR21 pKa = 11.84MHH23 pKa = 6.68KK24 pKa = 10.49ANRR27 pKa = 11.84TPRR30 pKa = 11.84FMRR33 pKa = 11.84GGIRR37 pKa = 11.84LL38 pKa = 3.51
Molecular weight: 4.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1386 |
38 |
547 |
231.0 |
26.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.215 ± 1.629 | 1.227 ± 0.42 |
5.195 ± 0.479 | 8.081 ± 1.214 |
3.752 ± 0.77 | 5.411 ± 0.634 |
1.876 ± 0.295 | 5.628 ± 0.5 |
8.225 ± 2.436 | 6.205 ± 0.533 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.958 ± 0.306 | 6.349 ± 1.009 |
3.752 ± 0.776 | 4.69 ± 1.333 |
4.834 ± 1.219 | 6.205 ± 0.693 |
7.648 ± 1.416 | 4.618 ± 0.529 |
1.299 ± 0.219 | 4.834 ± 0.598 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
