
Firmicutes bacterium CAG:65
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
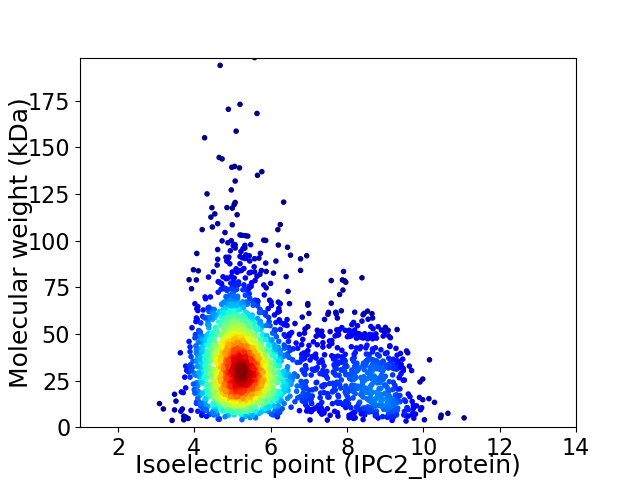
Virtual 2D-PAGE plot for 2512 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
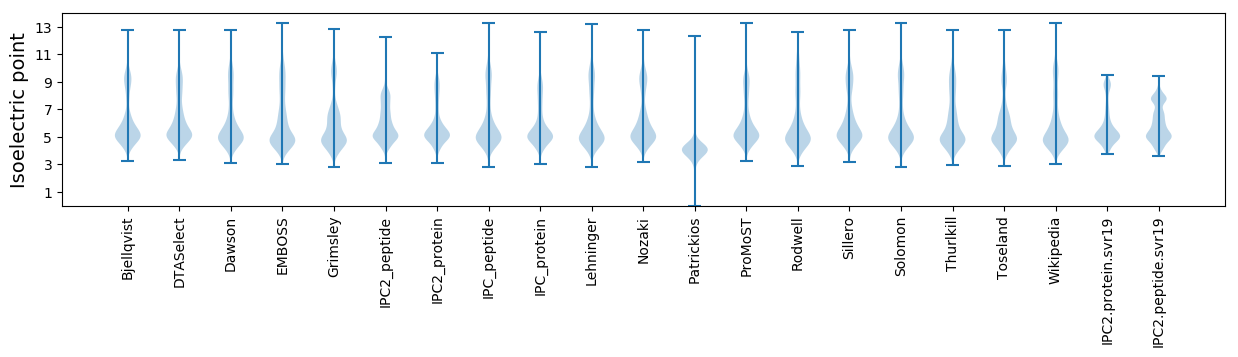
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6EJ78|R6EJ78_9FIRM GTP 3' 8-cyclase OS=Firmicutes bacterium CAG:65 OX=1262994 GN=moaA PE=3 SV=1
MM1 pKa = 7.55KK2 pKa = 10.48KK3 pKa = 10.11FLALALASVMAVSLVACGSTNADD26 pKa = 3.37TNAEE30 pKa = 3.99SSAAATTTAAATEE43 pKa = 4.42SGSEE47 pKa = 4.09EE48 pKa = 4.39PGLKK52 pKa = 9.55IAIVSSPSGVDD63 pKa = 3.24DD64 pKa = 5.18GSFNQDD70 pKa = 2.53NYY72 pKa = 11.46NGVLKK77 pKa = 10.55FIEE80 pKa = 4.56EE81 pKa = 4.22NPNATVTPIRR91 pKa = 11.84EE92 pKa = 4.18EE93 pKa = 4.24TGDD96 pKa = 3.44SAAAVQAVADD106 pKa = 4.0VVADD110 pKa = 3.57YY111 pKa = 11.33DD112 pKa = 4.73VIVCCGFQFAGIGNLAQEE130 pKa = 4.43NPDD133 pKa = 3.49TKK135 pKa = 10.83FILIDD140 pKa = 3.3SWPTIDD146 pKa = 3.79GTEE149 pKa = 3.88VTADD153 pKa = 3.36QVIDD157 pKa = 3.76NVYY160 pKa = 8.94AAYY163 pKa = 10.34YY164 pKa = 10.28AEE166 pKa = 4.23QEE168 pKa = 4.32SGFYY172 pKa = 10.71AGMAAALSTTTGKK185 pKa = 10.3VAVVNGIAYY194 pKa = 7.57PSNVNYY200 pKa = 10.29QYY202 pKa = 11.57GFEE205 pKa = 4.46CGVKK209 pKa = 9.87YY210 pKa = 11.28VNGTEE215 pKa = 4.03GMNVEE220 pKa = 4.58VVEE223 pKa = 4.41LPSYY227 pKa = 11.03AGTDD231 pKa = 3.29VTGANVGGNYY241 pKa = 9.74VGNFSDD247 pKa = 3.74EE248 pKa = 4.15ATGKK252 pKa = 10.29VLGNALIAEE261 pKa = 4.61GVDD264 pKa = 3.11ILFVAAGGSGNGVFTAAKK282 pKa = 9.1EE283 pKa = 3.86ADD285 pKa = 3.6GVKK288 pKa = 10.64VIGCDD293 pKa = 2.9VDD295 pKa = 4.4QYY297 pKa = 12.01DD298 pKa = 4.75DD299 pKa = 4.02GANGDD304 pKa = 4.13SNIVLTSGLKK314 pKa = 9.85VMHH317 pKa = 6.38DD318 pKa = 3.26TVYY321 pKa = 11.23NVLNEE326 pKa = 4.2VKK328 pKa = 10.55DD329 pKa = 4.28GSFKK333 pKa = 10.7GANVTLTAADD343 pKa = 4.09DD344 pKa = 3.63ATGFVSEE351 pKa = 4.93EE352 pKa = 4.2GRR354 pKa = 11.84CQLTAEE360 pKa = 4.91AIEE363 pKa = 5.01KK364 pKa = 10.37INAAQALLKK373 pKa = 10.15EE374 pKa = 4.6GKK376 pKa = 9.2IVPAANFNGVTPEE389 pKa = 3.9TFTWW393 pKa = 3.41
MM1 pKa = 7.55KK2 pKa = 10.48KK3 pKa = 10.11FLALALASVMAVSLVACGSTNADD26 pKa = 3.37TNAEE30 pKa = 3.99SSAAATTTAAATEE43 pKa = 4.42SGSEE47 pKa = 4.09EE48 pKa = 4.39PGLKK52 pKa = 9.55IAIVSSPSGVDD63 pKa = 3.24DD64 pKa = 5.18GSFNQDD70 pKa = 2.53NYY72 pKa = 11.46NGVLKK77 pKa = 10.55FIEE80 pKa = 4.56EE81 pKa = 4.22NPNATVTPIRR91 pKa = 11.84EE92 pKa = 4.18EE93 pKa = 4.24TGDD96 pKa = 3.44SAAAVQAVADD106 pKa = 4.0VVADD110 pKa = 3.57YY111 pKa = 11.33DD112 pKa = 4.73VIVCCGFQFAGIGNLAQEE130 pKa = 4.43NPDD133 pKa = 3.49TKK135 pKa = 10.83FILIDD140 pKa = 3.3SWPTIDD146 pKa = 3.79GTEE149 pKa = 3.88VTADD153 pKa = 3.36QVIDD157 pKa = 3.76NVYY160 pKa = 8.94AAYY163 pKa = 10.34YY164 pKa = 10.28AEE166 pKa = 4.23QEE168 pKa = 4.32SGFYY172 pKa = 10.71AGMAAALSTTTGKK185 pKa = 10.3VAVVNGIAYY194 pKa = 7.57PSNVNYY200 pKa = 10.29QYY202 pKa = 11.57GFEE205 pKa = 4.46CGVKK209 pKa = 9.87YY210 pKa = 11.28VNGTEE215 pKa = 4.03GMNVEE220 pKa = 4.58VVEE223 pKa = 4.41LPSYY227 pKa = 11.03AGTDD231 pKa = 3.29VTGANVGGNYY241 pKa = 9.74VGNFSDD247 pKa = 3.74EE248 pKa = 4.15ATGKK252 pKa = 10.29VLGNALIAEE261 pKa = 4.61GVDD264 pKa = 3.11ILFVAAGGSGNGVFTAAKK282 pKa = 9.1EE283 pKa = 3.86ADD285 pKa = 3.6GVKK288 pKa = 10.64VIGCDD293 pKa = 2.9VDD295 pKa = 4.4QYY297 pKa = 12.01DD298 pKa = 4.75DD299 pKa = 4.02GANGDD304 pKa = 4.13SNIVLTSGLKK314 pKa = 9.85VMHH317 pKa = 6.38DD318 pKa = 3.26TVYY321 pKa = 11.23NVLNEE326 pKa = 4.2VKK328 pKa = 10.55DD329 pKa = 4.28GSFKK333 pKa = 10.7GANVTLTAADD343 pKa = 4.09DD344 pKa = 3.63ATGFVSEE351 pKa = 4.93EE352 pKa = 4.2GRR354 pKa = 11.84CQLTAEE360 pKa = 4.91AIEE363 pKa = 5.01KK364 pKa = 10.37INAAQALLKK373 pKa = 10.15EE374 pKa = 4.6GKK376 pKa = 9.2IVPAANFNGVTPEE389 pKa = 3.9TFTWW393 pKa = 3.41
Molecular weight: 40.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6F6X0|R6F6X0_9FIRM Cyclic nucleotide-binding domain protein OS=Firmicutes bacterium CAG:65 OX=1262994 GN=BN749_01356 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.36KK9 pKa = 7.93RR10 pKa = 11.84SHH12 pKa = 6.15SKK14 pKa = 8.59VHH16 pKa = 6.03GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.36KK9 pKa = 7.93RR10 pKa = 11.84SHH12 pKa = 6.15SKK14 pKa = 8.59VHH16 pKa = 6.03GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 4.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
798008 |
31 |
1739 |
317.7 |
35.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.765 ± 0.047 | 1.498 ± 0.019 |
5.632 ± 0.04 | 7.663 ± 0.062 |
3.965 ± 0.039 | 7.17 ± 0.04 |
1.772 ± 0.024 | 7.106 ± 0.042 |
6.224 ± 0.043 | 9.19 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.151 ± 0.027 | 4.219 ± 0.034 |
3.321 ± 0.028 | 3.465 ± 0.031 |
4.453 ± 0.041 | 5.508 ± 0.038 |
5.654 ± 0.035 | 7.074 ± 0.04 |
0.926 ± 0.018 | 4.241 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
