
Avian paramyxovirus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Paraavulavirus; Avian paraavulavirus 4
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
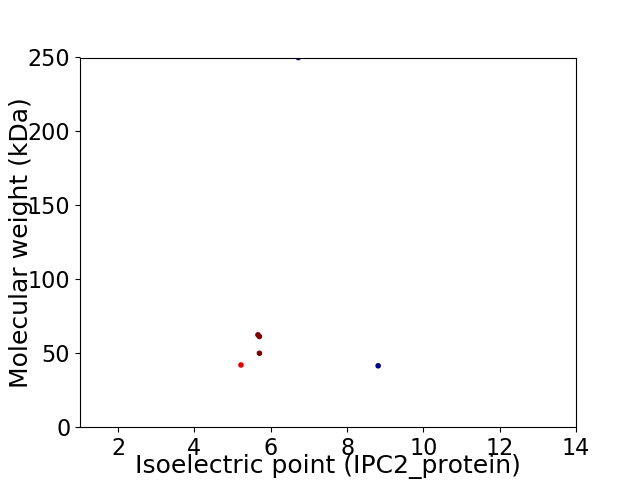
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G5D6F8|G5D6F8_9MONO Matrix protein OS=Avian paramyxovirus 4 OX=28274 GN=M PE=4 SV=1
MM1 pKa = 8.2DD2 pKa = 4.28FTDD5 pKa = 3.49IDD7 pKa = 3.99AVNSLIEE14 pKa = 4.2SSSAIIDD21 pKa = 4.24SIQHH25 pKa = 6.27GGLQPSGTVGLSQIPKK41 pKa = 9.85GITSALTKK49 pKa = 10.3AWEE52 pKa = 4.33AEE54 pKa = 3.8AATAGNGDD62 pKa = 4.04TQHH65 pKa = 6.92KK66 pKa = 9.11PDD68 pKa = 4.52SPEE71 pKa = 3.61DD72 pKa = 3.57HH73 pKa = 6.59QANDD77 pKa = 3.54TDD79 pKa = 4.21SPEE82 pKa = 4.12DD83 pKa = 3.16TGTNQTIQEE92 pKa = 4.19ANIVEE97 pKa = 4.59TPHH100 pKa = 7.17PEE102 pKa = 3.86VLSAAKK108 pKa = 10.28ARR110 pKa = 11.84LKK112 pKa = 10.52RR113 pKa = 11.84PKK115 pKa = 10.13AGRR118 pKa = 11.84DD119 pKa = 3.15THH121 pKa = 7.16DD122 pKa = 4.34NPSAQPDD129 pKa = 3.72HH130 pKa = 6.39FLKK133 pKa = 10.8GGPLSPQPAAPWVQSPPIHH152 pKa = 6.48GGPGTVDD159 pKa = 3.27PRR161 pKa = 11.84PSQTQDD167 pKa = 2.49HH168 pKa = 6.64SLTGEE173 pKa = 3.94KK174 pKa = 9.01WQSSPTKK181 pKa = 10.37QPEE184 pKa = 4.07TLNWWNGATRR194 pKa = 11.84GAPQSEE200 pKa = 4.45LNQPDD205 pKa = 3.62STVYY209 pKa = 11.23ADD211 pKa = 3.76TAPPSAASACMTTDD225 pKa = 3.51QVQLLMKK232 pKa = 9.77EE233 pKa = 4.25VADD236 pKa = 3.95MKK238 pKa = 11.11SLLQALVKK246 pKa = 10.54NLAVLPQLRR255 pKa = 11.84NEE257 pKa = 4.05VAAIRR262 pKa = 11.84TSQAMIEE269 pKa = 4.22GTLNSIKK276 pKa = 10.48ILDD279 pKa = 4.05PGNYY283 pKa = 9.02QEE285 pKa = 5.53SSLNSWFKK293 pKa = 10.31PRR295 pKa = 11.84QDD297 pKa = 3.2HH298 pKa = 5.93AVVVSGPGNPLTMPTPIQDD317 pKa = 3.28NTIFLDD323 pKa = 3.7EE324 pKa = 4.7LARR327 pKa = 11.84PHH329 pKa = 6.98PSLVNPSPPTTNTNVDD345 pKa = 4.43LGPQKK350 pKa = 10.25QAAIAYY356 pKa = 9.21ISAKK360 pKa = 10.48CKK362 pKa = 10.78DD363 pKa = 3.41PGKK366 pKa = 10.12RR367 pKa = 11.84DD368 pKa = 3.54QLSKK372 pKa = 10.72LIEE375 pKa = 4.23RR376 pKa = 11.84ATTLSEE382 pKa = 3.71INKK385 pKa = 9.41VKK387 pKa = 10.56RR388 pKa = 11.84QALGLL393 pKa = 3.93
MM1 pKa = 8.2DD2 pKa = 4.28FTDD5 pKa = 3.49IDD7 pKa = 3.99AVNSLIEE14 pKa = 4.2SSSAIIDD21 pKa = 4.24SIQHH25 pKa = 6.27GGLQPSGTVGLSQIPKK41 pKa = 9.85GITSALTKK49 pKa = 10.3AWEE52 pKa = 4.33AEE54 pKa = 3.8AATAGNGDD62 pKa = 4.04TQHH65 pKa = 6.92KK66 pKa = 9.11PDD68 pKa = 4.52SPEE71 pKa = 3.61DD72 pKa = 3.57HH73 pKa = 6.59QANDD77 pKa = 3.54TDD79 pKa = 4.21SPEE82 pKa = 4.12DD83 pKa = 3.16TGTNQTIQEE92 pKa = 4.19ANIVEE97 pKa = 4.59TPHH100 pKa = 7.17PEE102 pKa = 3.86VLSAAKK108 pKa = 10.28ARR110 pKa = 11.84LKK112 pKa = 10.52RR113 pKa = 11.84PKK115 pKa = 10.13AGRR118 pKa = 11.84DD119 pKa = 3.15THH121 pKa = 7.16DD122 pKa = 4.34NPSAQPDD129 pKa = 3.72HH130 pKa = 6.39FLKK133 pKa = 10.8GGPLSPQPAAPWVQSPPIHH152 pKa = 6.48GGPGTVDD159 pKa = 3.27PRR161 pKa = 11.84PSQTQDD167 pKa = 2.49HH168 pKa = 6.64SLTGEE173 pKa = 3.94KK174 pKa = 9.01WQSSPTKK181 pKa = 10.37QPEE184 pKa = 4.07TLNWWNGATRR194 pKa = 11.84GAPQSEE200 pKa = 4.45LNQPDD205 pKa = 3.62STVYY209 pKa = 11.23ADD211 pKa = 3.76TAPPSAASACMTTDD225 pKa = 3.51QVQLLMKK232 pKa = 9.77EE233 pKa = 4.25VADD236 pKa = 3.95MKK238 pKa = 11.11SLLQALVKK246 pKa = 10.54NLAVLPQLRR255 pKa = 11.84NEE257 pKa = 4.05VAAIRR262 pKa = 11.84TSQAMIEE269 pKa = 4.22GTLNSIKK276 pKa = 10.48ILDD279 pKa = 4.05PGNYY283 pKa = 9.02QEE285 pKa = 5.53SSLNSWFKK293 pKa = 10.31PRR295 pKa = 11.84QDD297 pKa = 3.2HH298 pKa = 5.93AVVVSGPGNPLTMPTPIQDD317 pKa = 3.28NTIFLDD323 pKa = 3.7EE324 pKa = 4.7LARR327 pKa = 11.84PHH329 pKa = 6.98PSLVNPSPPTTNTNVDD345 pKa = 4.43LGPQKK350 pKa = 10.25QAAIAYY356 pKa = 9.21ISAKK360 pKa = 10.48CKK362 pKa = 10.78DD363 pKa = 3.41PGKK366 pKa = 10.12RR367 pKa = 11.84DD368 pKa = 3.54QLSKK372 pKa = 10.72LIEE375 pKa = 4.23RR376 pKa = 11.84ATTLSEE382 pKa = 3.71INKK385 pKa = 9.41VKK387 pKa = 10.56RR388 pKa = 11.84QALGLL393 pKa = 3.93
Molecular weight: 42.05 kDa
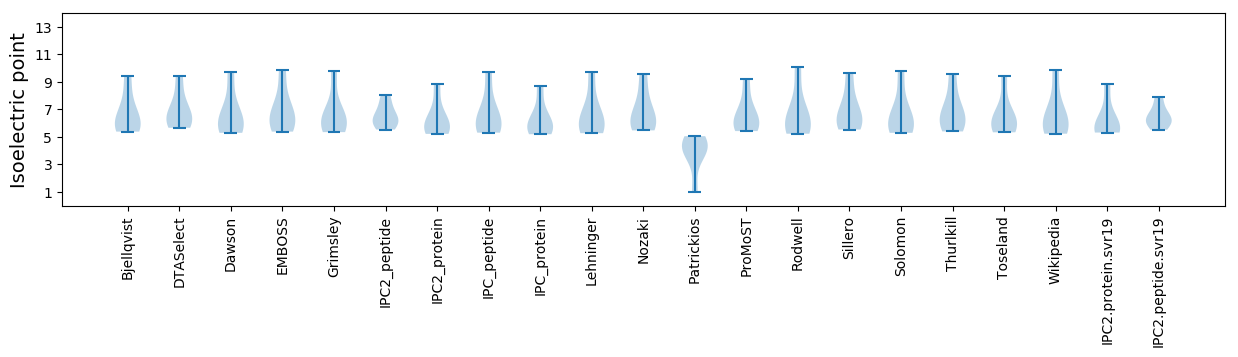
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G5D6F9|G5D6F9_9MONO Fusion glycoprotein F0 OS=Avian paramyxovirus 4 OX=28274 GN=F PE=3 SV=1
MM1 pKa = 7.74ADD3 pKa = 3.45MDD5 pKa = 4.25TVYY8 pKa = 10.71INLMADD14 pKa = 3.69DD15 pKa = 4.42PTHH18 pKa = 6.18QKK20 pKa = 10.6EE21 pKa = 4.43LLSFPLVPVTGPDD34 pKa = 3.39GKK36 pKa = 10.78KK37 pKa = 9.81EE38 pKa = 3.91LQHH41 pKa = 5.62QVRR44 pKa = 11.84TQSLLASDD52 pKa = 3.93KK53 pKa = 9.01QTEE56 pKa = 4.07RR57 pKa = 11.84FIFLNTYY64 pKa = 9.8GFIYY68 pKa = 9.16DD69 pKa = 3.76TTPDD73 pKa = 3.29KK74 pKa = 9.65TTFSTPEE81 pKa = 4.67HH82 pKa = 6.29INQPKK87 pKa = 8.47RR88 pKa = 11.84TMVSAAMMTIGLVPANIPLNEE109 pKa = 3.88LTATVFGLKK118 pKa = 10.05VRR120 pKa = 11.84VRR122 pKa = 11.84KK123 pKa = 8.75SARR126 pKa = 11.84YY127 pKa = 9.39RR128 pKa = 11.84EE129 pKa = 4.36VVWYY133 pKa = 9.53QCNPVPALLAATRR146 pKa = 11.84FGRR149 pKa = 11.84QGGLEE154 pKa = 4.13SSTGVSVKK162 pKa = 10.51APEE165 pKa = 5.52KK166 pKa = 10.09IDD168 pKa = 3.52CEE170 pKa = 3.95KK171 pKa = 11.01DD172 pKa = 3.21YY173 pKa = 11.09TYY175 pKa = 11.38YY176 pKa = 10.37PYY178 pKa = 10.5FLSVCYY184 pKa = 10.01IATSNLFKK192 pKa = 10.5VPKK195 pKa = 9.4MVANATNSQLYY206 pKa = 9.02HH207 pKa = 6.19LTMQVTFAFPKK218 pKa = 10.26NIPPANQKK226 pKa = 10.5LLTQVDD232 pKa = 3.83EE233 pKa = 4.71GFEE236 pKa = 4.29GTVDD240 pKa = 3.43CHH242 pKa = 6.74FGNMLKK248 pKa = 10.04KK249 pKa = 10.15DD250 pKa = 3.48RR251 pKa = 11.84KK252 pKa = 10.08GNMRR256 pKa = 11.84TLSQAADD263 pKa = 3.29KK264 pKa = 10.17VRR266 pKa = 11.84RR267 pKa = 11.84MNILVGIFDD276 pKa = 3.87LHH278 pKa = 6.63GPTLFLEE285 pKa = 4.56YY286 pKa = 9.93TGKK289 pKa = 8.51LTKK292 pKa = 10.53ALLGFMSTSRR302 pKa = 11.84TAIIPISQLNPMLGQLMWSSDD323 pKa = 3.25AQIVKK328 pKa = 10.1LRR330 pKa = 11.84VVITTSKK337 pKa = 10.43RR338 pKa = 11.84GPCGGEE344 pKa = 3.48QEE346 pKa = 4.66YY347 pKa = 11.4VLDD350 pKa = 4.31PKK352 pKa = 10.51FTVKK356 pKa = 10.42KK357 pKa = 9.94EE358 pKa = 3.9KK359 pKa = 10.68ARR361 pKa = 11.84LNPFKK366 pKa = 10.7KK367 pKa = 10.21AAQQ370 pKa = 3.49
MM1 pKa = 7.74ADD3 pKa = 3.45MDD5 pKa = 4.25TVYY8 pKa = 10.71INLMADD14 pKa = 3.69DD15 pKa = 4.42PTHH18 pKa = 6.18QKK20 pKa = 10.6EE21 pKa = 4.43LLSFPLVPVTGPDD34 pKa = 3.39GKK36 pKa = 10.78KK37 pKa = 9.81EE38 pKa = 3.91LQHH41 pKa = 5.62QVRR44 pKa = 11.84TQSLLASDD52 pKa = 3.93KK53 pKa = 9.01QTEE56 pKa = 4.07RR57 pKa = 11.84FIFLNTYY64 pKa = 9.8GFIYY68 pKa = 9.16DD69 pKa = 3.76TTPDD73 pKa = 3.29KK74 pKa = 9.65TTFSTPEE81 pKa = 4.67HH82 pKa = 6.29INQPKK87 pKa = 8.47RR88 pKa = 11.84TMVSAAMMTIGLVPANIPLNEE109 pKa = 3.88LTATVFGLKK118 pKa = 10.05VRR120 pKa = 11.84VRR122 pKa = 11.84KK123 pKa = 8.75SARR126 pKa = 11.84YY127 pKa = 9.39RR128 pKa = 11.84EE129 pKa = 4.36VVWYY133 pKa = 9.53QCNPVPALLAATRR146 pKa = 11.84FGRR149 pKa = 11.84QGGLEE154 pKa = 4.13SSTGVSVKK162 pKa = 10.51APEE165 pKa = 5.52KK166 pKa = 10.09IDD168 pKa = 3.52CEE170 pKa = 3.95KK171 pKa = 11.01DD172 pKa = 3.21YY173 pKa = 11.09TYY175 pKa = 11.38YY176 pKa = 10.37PYY178 pKa = 10.5FLSVCYY184 pKa = 10.01IATSNLFKK192 pKa = 10.5VPKK195 pKa = 9.4MVANATNSQLYY206 pKa = 9.02HH207 pKa = 6.19LTMQVTFAFPKK218 pKa = 10.26NIPPANQKK226 pKa = 10.5LLTQVDD232 pKa = 3.83EE233 pKa = 4.71GFEE236 pKa = 4.29GTVDD240 pKa = 3.43CHH242 pKa = 6.74FGNMLKK248 pKa = 10.04KK249 pKa = 10.15DD250 pKa = 3.48RR251 pKa = 11.84KK252 pKa = 10.08GNMRR256 pKa = 11.84TLSQAADD263 pKa = 3.29KK264 pKa = 10.17VRR266 pKa = 11.84RR267 pKa = 11.84MNILVGIFDD276 pKa = 3.87LHH278 pKa = 6.63GPTLFLEE285 pKa = 4.56YY286 pKa = 9.93TGKK289 pKa = 8.51LTKK292 pKa = 10.53ALLGFMSTSRR302 pKa = 11.84TAIIPISQLNPMLGQLMWSSDD323 pKa = 3.25AQIVKK328 pKa = 10.1LRR330 pKa = 11.84VVITTSKK337 pKa = 10.43RR338 pKa = 11.84GPCGGEE344 pKa = 3.48QEE346 pKa = 4.66YY347 pKa = 11.4VLDD350 pKa = 4.31PKK352 pKa = 10.51FTVKK356 pKa = 10.42KK357 pKa = 9.94EE358 pKa = 3.9KK359 pKa = 10.68ARR361 pKa = 11.84LNPFKK366 pKa = 10.7KK367 pKa = 10.21AAQQ370 pKa = 3.49
Molecular weight: 41.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4562 |
370 |
2211 |
760.3 |
84.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.278 ± 0.849 | 1.797 ± 0.328 |
5.151 ± 0.235 | 4.472 ± 0.369 |
3.31 ± 0.393 | 5.612 ± 0.352 |
2.148 ± 0.419 | 6.094 ± 0.466 |
4.143 ± 0.616 | 11.201 ± 0.799 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.302 ± 0.28 | 4.822 ± 0.35 |
5.217 ± 0.595 | 5.129 ± 0.421 |
5.261 ± 0.353 | 8.593 ± 0.628 |
6.664 ± 0.547 | 6.203 ± 0.399 |
1.162 ± 0.23 | 3.42 ± 0.425 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
