
Avian metaavulavirus 6
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Metaavulavirus
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
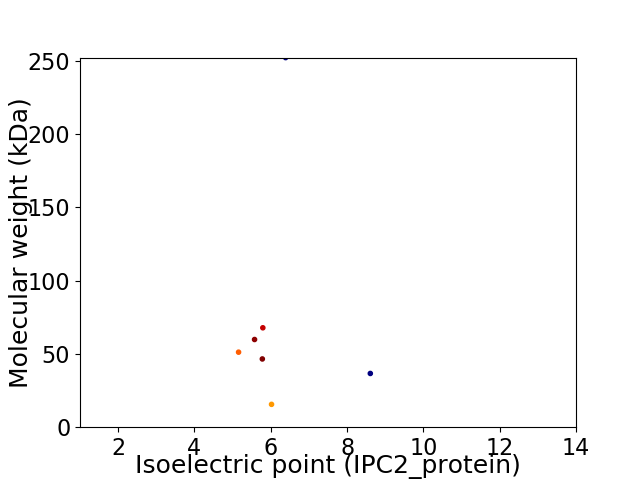
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|G5D6G3|G5D6G3_9MONO Phosphoprotein OS=Avian metaavulavirus 6 OX=2560316 GN=P PE=4 SV=1
MM1 pKa = 7.42SSVFTDD7 pKa = 3.69YY8 pKa = 11.8AKK10 pKa = 11.09LQDD13 pKa = 4.02ALVAPSKK20 pKa = 10.52RR21 pKa = 11.84KK22 pKa = 9.69VDD24 pKa = 3.77SAPSGLLRR32 pKa = 11.84VGIPVCVLLSEE43 pKa = 4.89DD44 pKa = 3.83PEE46 pKa = 4.29EE47 pKa = 4.06RR48 pKa = 11.84WGFVCFCMRR57 pKa = 11.84WVVSDD62 pKa = 3.69SATEE66 pKa = 3.88AMRR69 pKa = 11.84VGAMLSILSAHH80 pKa = 6.81ASNMRR85 pKa = 11.84SHH87 pKa = 6.44VALAARR93 pKa = 11.84CGDD96 pKa = 3.3ADD98 pKa = 3.76INILEE103 pKa = 4.38VEE105 pKa = 5.02AIDD108 pKa = 5.02HH109 pKa = 5.5QNQTIRR115 pKa = 11.84FTGRR119 pKa = 11.84SNVTDD124 pKa = 3.37GRR126 pKa = 11.84ARR128 pKa = 11.84QMYY131 pKa = 10.38AIAQDD136 pKa = 4.5LPPSYY141 pKa = 11.42NNGSPFVNRR150 pKa = 11.84DD151 pKa = 2.78IEE153 pKa = 4.84DD154 pKa = 3.77NYY156 pKa = 8.75PTDD159 pKa = 3.45MSEE162 pKa = 4.5LLNMVYY168 pKa = 10.38SVATQIWVAAMKK180 pKa = 10.93SMTAPDD186 pKa = 3.71TSSEE190 pKa = 4.37SEE192 pKa = 3.84GRR194 pKa = 11.84RR195 pKa = 11.84LAKK198 pKa = 10.21YY199 pKa = 8.56IQQNRR204 pKa = 11.84VIRR207 pKa = 11.84STILAPATRR216 pKa = 11.84GEE218 pKa = 4.26CTRR221 pKa = 11.84IIRR224 pKa = 11.84SSLVIRR230 pKa = 11.84HH231 pKa = 5.71FLITEE236 pKa = 4.4IKK238 pKa = 9.89RR239 pKa = 11.84ATSMGSNTTRR249 pKa = 11.84YY250 pKa = 8.83YY251 pKa = 10.73ATVGDD256 pKa = 3.58AAAYY260 pKa = 7.74FKK262 pKa = 10.82NAGMAAFFLTLRR274 pKa = 11.84FGIGTKK280 pKa = 10.51YY281 pKa = 9.43STLAVSALSADD292 pKa = 3.73MKK294 pKa = 10.77KK295 pKa = 9.96LQSLIRR301 pKa = 11.84VYY303 pKa = 10.2QSKK306 pKa = 11.07GEE308 pKa = 4.34DD309 pKa = 3.65GPYY312 pKa = 9.38MAFLEE317 pKa = 5.16DD318 pKa = 3.68SDD320 pKa = 4.92LMSFAPGNYY329 pKa = 8.64PLMYY333 pKa = 9.44SYY335 pKa = 11.87AMGVGSILEE344 pKa = 3.99ASIARR349 pKa = 11.84YY350 pKa = 9.26QFARR354 pKa = 11.84SFMNDD359 pKa = 2.11TFYY362 pKa = 11.54RR363 pKa = 11.84LGVEE367 pKa = 4.17TAQRR371 pKa = 11.84NQGSLDD377 pKa = 3.5EE378 pKa = 4.61NLAKK382 pKa = 10.28EE383 pKa = 4.22LQLSGAEE390 pKa = 3.53RR391 pKa = 11.84RR392 pKa = 11.84AVQEE396 pKa = 4.23LVTSLDD402 pKa = 3.73LAGEE406 pKa = 4.17APVPQRR412 pKa = 11.84QPTFLNDD419 pKa = 3.12QEE421 pKa = 5.41YY422 pKa = 10.85EE423 pKa = 4.05DD424 pKa = 5.44DD425 pKa = 4.08PPARR429 pKa = 11.84RR430 pKa = 11.84QRR432 pKa = 11.84IEE434 pKa = 4.36DD435 pKa = 3.89TPDD438 pKa = 3.91DD439 pKa = 4.98DD440 pKa = 5.52GASQAPPTPGAGLTPYY456 pKa = 10.59SDD458 pKa = 3.45NASGLDD464 pKa = 3.3II465 pKa = 5.27
MM1 pKa = 7.42SSVFTDD7 pKa = 3.69YY8 pKa = 11.8AKK10 pKa = 11.09LQDD13 pKa = 4.02ALVAPSKK20 pKa = 10.52RR21 pKa = 11.84KK22 pKa = 9.69VDD24 pKa = 3.77SAPSGLLRR32 pKa = 11.84VGIPVCVLLSEE43 pKa = 4.89DD44 pKa = 3.83PEE46 pKa = 4.29EE47 pKa = 4.06RR48 pKa = 11.84WGFVCFCMRR57 pKa = 11.84WVVSDD62 pKa = 3.69SATEE66 pKa = 3.88AMRR69 pKa = 11.84VGAMLSILSAHH80 pKa = 6.81ASNMRR85 pKa = 11.84SHH87 pKa = 6.44VALAARR93 pKa = 11.84CGDD96 pKa = 3.3ADD98 pKa = 3.76INILEE103 pKa = 4.38VEE105 pKa = 5.02AIDD108 pKa = 5.02HH109 pKa = 5.5QNQTIRR115 pKa = 11.84FTGRR119 pKa = 11.84SNVTDD124 pKa = 3.37GRR126 pKa = 11.84ARR128 pKa = 11.84QMYY131 pKa = 10.38AIAQDD136 pKa = 4.5LPPSYY141 pKa = 11.42NNGSPFVNRR150 pKa = 11.84DD151 pKa = 2.78IEE153 pKa = 4.84DD154 pKa = 3.77NYY156 pKa = 8.75PTDD159 pKa = 3.45MSEE162 pKa = 4.5LLNMVYY168 pKa = 10.38SVATQIWVAAMKK180 pKa = 10.93SMTAPDD186 pKa = 3.71TSSEE190 pKa = 4.37SEE192 pKa = 3.84GRR194 pKa = 11.84RR195 pKa = 11.84LAKK198 pKa = 10.21YY199 pKa = 8.56IQQNRR204 pKa = 11.84VIRR207 pKa = 11.84STILAPATRR216 pKa = 11.84GEE218 pKa = 4.26CTRR221 pKa = 11.84IIRR224 pKa = 11.84SSLVIRR230 pKa = 11.84HH231 pKa = 5.71FLITEE236 pKa = 4.4IKK238 pKa = 9.89RR239 pKa = 11.84ATSMGSNTTRR249 pKa = 11.84YY250 pKa = 8.83YY251 pKa = 10.73ATVGDD256 pKa = 3.58AAAYY260 pKa = 7.74FKK262 pKa = 10.82NAGMAAFFLTLRR274 pKa = 11.84FGIGTKK280 pKa = 10.51YY281 pKa = 9.43STLAVSALSADD292 pKa = 3.73MKK294 pKa = 10.77KK295 pKa = 9.96LQSLIRR301 pKa = 11.84VYY303 pKa = 10.2QSKK306 pKa = 11.07GEE308 pKa = 4.34DD309 pKa = 3.65GPYY312 pKa = 9.38MAFLEE317 pKa = 5.16DD318 pKa = 3.68SDD320 pKa = 4.92LMSFAPGNYY329 pKa = 8.64PLMYY333 pKa = 9.44SYY335 pKa = 11.87AMGVGSILEE344 pKa = 3.99ASIARR349 pKa = 11.84YY350 pKa = 9.26QFARR354 pKa = 11.84SFMNDD359 pKa = 2.11TFYY362 pKa = 11.54RR363 pKa = 11.84LGVEE367 pKa = 4.17TAQRR371 pKa = 11.84NQGSLDD377 pKa = 3.5EE378 pKa = 4.61NLAKK382 pKa = 10.28EE383 pKa = 4.22LQLSGAEE390 pKa = 3.53RR391 pKa = 11.84RR392 pKa = 11.84AVQEE396 pKa = 4.23LVTSLDD402 pKa = 3.73LAGEE406 pKa = 4.17APVPQRR412 pKa = 11.84QPTFLNDD419 pKa = 3.12QEE421 pKa = 5.41YY422 pKa = 10.85EE423 pKa = 4.05DD424 pKa = 5.44DD425 pKa = 4.08PPARR429 pKa = 11.84RR430 pKa = 11.84QRR432 pKa = 11.84IEE434 pKa = 4.36DD435 pKa = 3.89TPDD438 pKa = 3.91DD439 pKa = 4.98DD440 pKa = 5.52GASQAPPTPGAGLTPYY456 pKa = 10.59SDD458 pKa = 3.45NASGLDD464 pKa = 3.3II465 pKa = 5.27
Molecular weight: 51.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G5D6G5|G5D6G5_9MONO Fusion glycoprotein F0 OS=Avian metaavulavirus 6 OX=2560316 GN=F PE=3 SV=1
MM1 pKa = 7.19ATXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXLQPQLRR41 pKa = 11.84TQYY44 pKa = 11.0LGDD47 pKa = 3.37VTGGKK52 pKa = 9.84KK53 pKa = 9.8SAIFVNCYY61 pKa = 10.56GFVEE65 pKa = 4.29DD66 pKa = 4.99HH67 pKa = 6.8GGRR70 pKa = 11.84DD71 pKa = 3.4SGFSPISEE79 pKa = 4.15EE80 pKa = 4.5SKK82 pKa = 10.92GSTVTAACITLGSIEE97 pKa = 4.36YY98 pKa = 10.47DD99 pKa = 3.18SDD101 pKa = 3.18IKK103 pKa = 10.93EE104 pKa = 4.22VAKK107 pKa = 10.58ACYY110 pKa = 9.07NLQVSVRR117 pKa = 11.84MSADD121 pKa = 2.95STQKK125 pKa = 10.21VVYY128 pKa = 8.98TINAKK133 pKa = 9.6PALLFSSRR141 pKa = 11.84VVRR144 pKa = 11.84AGGCVVAAEE153 pKa = 4.45GAIKK157 pKa = 10.41CPEE160 pKa = 3.99KK161 pKa = 9.16MTSDD165 pKa = 3.15RR166 pKa = 11.84LYY168 pKa = 9.74KK169 pKa = 10.02FRR171 pKa = 11.84VMFVSLTFLHH181 pKa = 6.75RR182 pKa = 11.84SSLFKK187 pKa = 10.81VSRR190 pKa = 11.84TVLSMRR196 pKa = 11.84NSALIAVQAEE206 pKa = 4.41VKK208 pKa = 10.66LGFDD212 pKa = 4.54LPLDD216 pKa = 3.87HH217 pKa = 7.56PMAKK221 pKa = 10.07YY222 pKa = 10.32LSKK225 pKa = 11.13EE226 pKa = 4.03DD227 pKa = 3.58GQLFATVWVHH237 pKa = 5.99LCNFKK242 pKa = 9.97RR243 pKa = 11.84TDD245 pKa = 3.0RR246 pKa = 11.84RR247 pKa = 11.84GVDD250 pKa = 2.93RR251 pKa = 11.84SVEE254 pKa = 4.03NIRR257 pKa = 11.84NKK259 pKa = 9.07VRR261 pKa = 11.84AMGLKK266 pKa = 8.96LTLCDD271 pKa = 3.23LWGPTLVCEE280 pKa = 4.65ATGKK284 pKa = 8.15MSKK287 pKa = 10.14YY288 pKa = 10.12ALGFFSEE295 pKa = 4.78TKK297 pKa = 10.42VGCHH301 pKa = 6.72PIWKK305 pKa = 9.49CNSTVAKK312 pKa = 10.45IMWSCTTWIASAKK325 pKa = 10.37AIIQASSARR334 pKa = 11.84ALLTSEE340 pKa = 5.43DD341 pKa = 3.89IEE343 pKa = 4.49AKK345 pKa = 10.52GAISTDD351 pKa = 3.02KK352 pKa = 11.09KK353 pKa = 10.19KK354 pKa = 9.38TDD356 pKa = 3.42GFNPFIKK363 pKa = 9.44TAKK366 pKa = 9.97
MM1 pKa = 7.19ATXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXLQPQLRR41 pKa = 11.84TQYY44 pKa = 11.0LGDD47 pKa = 3.37VTGGKK52 pKa = 9.84KK53 pKa = 9.8SAIFVNCYY61 pKa = 10.56GFVEE65 pKa = 4.29DD66 pKa = 4.99HH67 pKa = 6.8GGRR70 pKa = 11.84DD71 pKa = 3.4SGFSPISEE79 pKa = 4.15EE80 pKa = 4.5SKK82 pKa = 10.92GSTVTAACITLGSIEE97 pKa = 4.36YY98 pKa = 10.47DD99 pKa = 3.18SDD101 pKa = 3.18IKK103 pKa = 10.93EE104 pKa = 4.22VAKK107 pKa = 10.58ACYY110 pKa = 9.07NLQVSVRR117 pKa = 11.84MSADD121 pKa = 2.95STQKK125 pKa = 10.21VVYY128 pKa = 8.98TINAKK133 pKa = 9.6PALLFSSRR141 pKa = 11.84VVRR144 pKa = 11.84AGGCVVAAEE153 pKa = 4.45GAIKK157 pKa = 10.41CPEE160 pKa = 3.99KK161 pKa = 9.16MTSDD165 pKa = 3.15RR166 pKa = 11.84LYY168 pKa = 9.74KK169 pKa = 10.02FRR171 pKa = 11.84VMFVSLTFLHH181 pKa = 6.75RR182 pKa = 11.84SSLFKK187 pKa = 10.81VSRR190 pKa = 11.84TVLSMRR196 pKa = 11.84NSALIAVQAEE206 pKa = 4.41VKK208 pKa = 10.66LGFDD212 pKa = 4.54LPLDD216 pKa = 3.87HH217 pKa = 7.56PMAKK221 pKa = 10.07YY222 pKa = 10.32LSKK225 pKa = 11.13EE226 pKa = 4.03DD227 pKa = 3.58GQLFATVWVHH237 pKa = 5.99LCNFKK242 pKa = 9.97RR243 pKa = 11.84TDD245 pKa = 3.0RR246 pKa = 11.84RR247 pKa = 11.84GVDD250 pKa = 2.93RR251 pKa = 11.84SVEE254 pKa = 4.03NIRR257 pKa = 11.84NKK259 pKa = 9.07VRR261 pKa = 11.84AMGLKK266 pKa = 8.96LTLCDD271 pKa = 3.23LWGPTLVCEE280 pKa = 4.65ATGKK284 pKa = 8.15MSKK287 pKa = 10.14YY288 pKa = 10.12ALGFFSEE295 pKa = 4.78TKK297 pKa = 10.42VGCHH301 pKa = 6.72PIWKK305 pKa = 9.49CNSTVAKK312 pKa = 10.45IMWSCTTWIASAKK325 pKa = 10.37AIIQASSARR334 pKa = 11.84ALLTSEE340 pKa = 5.43DD341 pKa = 3.89IEE343 pKa = 4.49AKK345 pKa = 10.52GAISTDD351 pKa = 3.02KK352 pKa = 11.09KK353 pKa = 10.19KK354 pKa = 9.38TDD356 pKa = 3.42GFNPFIKK363 pKa = 9.44TAKK366 pKa = 9.97
Molecular weight: 36.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4812 |
142 |
2241 |
687.4 |
75.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.502 ± 0.869 | 2.057 ± 0.277 |
4.738 ± 0.348 | 5.403 ± 0.464 |
3.159 ± 0.373 | 6.068 ± 0.222 |
2.244 ± 0.459 | 6.796 ± 0.52 |
4.468 ± 0.502 | 10.287 ± 1.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.431 ± 0.242 | 4.426 ± 0.374 |
4.468 ± 0.338 | 4.281 ± 0.455 |
5.32 ± 0.455 | 8.084 ± 0.23 |
6.816 ± 0.494 | 6.671 ± 0.272 |
1.164 ± 0.238 | 2.951 ± 0.382 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
