
Bat Hp-betacoronavirus/Zhejiang2013
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Hibecovirus; Bat Hp-betacoronavirus Zhejiang2013
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
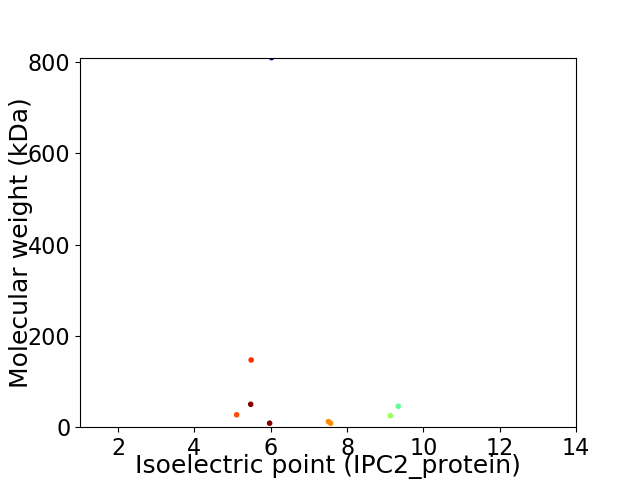
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
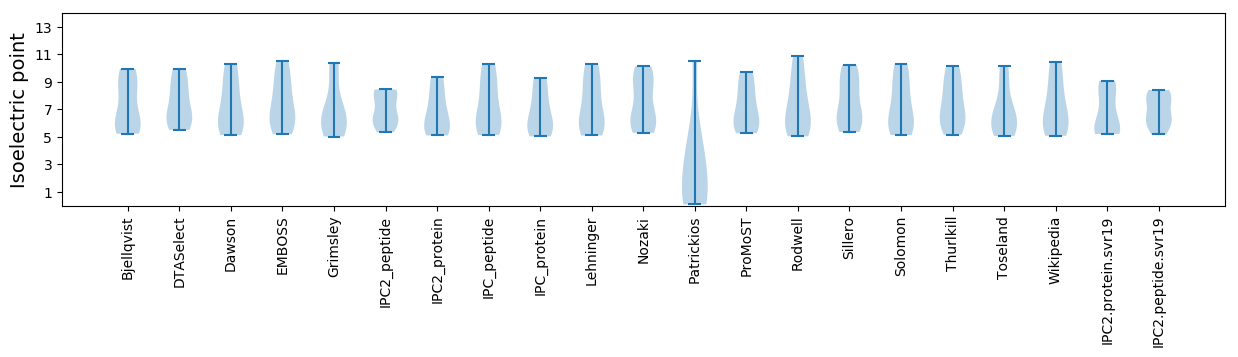
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A088DI26|A0A088DI26_9BETC Nucleoprotein OS=Bat Hp-betacoronavirus/Zhejiang2013 OX=1541205 PE=4 SV=1
MM1 pKa = 8.11DD2 pKa = 5.19FFSLYY7 pKa = 10.27TFGSFHH13 pKa = 5.26MHH15 pKa = 6.13AAKK18 pKa = 10.22LASLLSNSTEE28 pKa = 3.95HH29 pKa = 7.12VEE31 pKa = 4.89QAVQHH36 pKa = 6.5LNTKK40 pKa = 8.64LTADD44 pKa = 2.95IMLCIFLVIYY54 pKa = 7.4LICYY58 pKa = 8.01FLRR61 pKa = 11.84TDD63 pKa = 3.21SFIAVVFKK71 pKa = 10.85YY72 pKa = 10.42LAGLLTGGFLCLGLFLDD89 pKa = 4.96TPTLLLKK96 pKa = 10.03ATIGVVLFMFSLGFICRR113 pKa = 11.84ITLAIRR119 pKa = 11.84CKK121 pKa = 10.76SLVPLCADD129 pKa = 3.85DD130 pKa = 5.34DD131 pKa = 4.58CFVNYY136 pKa = 10.07NAGGKK141 pKa = 6.52TYY143 pKa = 10.82CMPFDD148 pKa = 4.5PNEE151 pKa = 4.25PYY153 pKa = 9.55LTLVVHH159 pKa = 6.4QNGITCGSYY168 pKa = 10.85KK169 pKa = 10.7LYY171 pKa = 10.91GDD173 pKa = 3.43VSIADD178 pKa = 4.61RR179 pKa = 11.84IYY181 pKa = 11.13LVTLTKK187 pKa = 10.32SVPYY191 pKa = 9.98SLQNIFDD198 pKa = 4.6AEE200 pKa = 4.3LCTIAFYY207 pKa = 10.48IADD210 pKa = 3.74CAVIEE215 pKa = 4.25DD216 pKa = 4.23HH217 pKa = 5.85TTAGKK222 pKa = 8.25TPRR225 pKa = 11.84LEE227 pKa = 4.47LKK229 pKa = 10.12SDD231 pKa = 4.79PIYY234 pKa = 10.19EE235 pKa = 4.47VPCATIDD242 pKa = 3.58VPLL245 pKa = 5.05
MM1 pKa = 8.11DD2 pKa = 5.19FFSLYY7 pKa = 10.27TFGSFHH13 pKa = 5.26MHH15 pKa = 6.13AAKK18 pKa = 10.22LASLLSNSTEE28 pKa = 3.95HH29 pKa = 7.12VEE31 pKa = 4.89QAVQHH36 pKa = 6.5LNTKK40 pKa = 8.64LTADD44 pKa = 2.95IMLCIFLVIYY54 pKa = 7.4LICYY58 pKa = 8.01FLRR61 pKa = 11.84TDD63 pKa = 3.21SFIAVVFKK71 pKa = 10.85YY72 pKa = 10.42LAGLLTGGFLCLGLFLDD89 pKa = 4.96TPTLLLKK96 pKa = 10.03ATIGVVLFMFSLGFICRR113 pKa = 11.84ITLAIRR119 pKa = 11.84CKK121 pKa = 10.76SLVPLCADD129 pKa = 3.85DD130 pKa = 5.34DD131 pKa = 4.58CFVNYY136 pKa = 10.07NAGGKK141 pKa = 6.52TYY143 pKa = 10.82CMPFDD148 pKa = 4.5PNEE151 pKa = 4.25PYY153 pKa = 9.55LTLVVHH159 pKa = 6.4QNGITCGSYY168 pKa = 10.85KK169 pKa = 10.7LYY171 pKa = 10.91GDD173 pKa = 3.43VSIADD178 pKa = 4.61RR179 pKa = 11.84IYY181 pKa = 11.13LVTLTKK187 pKa = 10.32SVPYY191 pKa = 9.98SLQNIFDD198 pKa = 4.6AEE200 pKa = 4.3LCTIAFYY207 pKa = 10.48IADD210 pKa = 3.74CAVIEE215 pKa = 4.25DD216 pKa = 4.23HH217 pKa = 5.85TTAGKK222 pKa = 8.25TPRR225 pKa = 11.84LEE227 pKa = 4.47LKK229 pKa = 10.12SDD231 pKa = 4.79PIYY234 pKa = 10.19EE235 pKa = 4.47VPCATIDD242 pKa = 3.58VPLL245 pKa = 5.05
Molecular weight: 27.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A088DIE1|A0A088DIE1_9BETC 2'-O-methyltransferase OS=Bat Hp-betacoronavirus/Zhejiang2013 OX=1541205 PE=3 SV=1
MM1 pKa = 7.31AAEE4 pKa = 4.32GQRR7 pKa = 11.84VTFLTQGSNQNNNNQEE23 pKa = 4.12NQDD26 pKa = 3.44GRR28 pKa = 11.84GARR31 pKa = 11.84PKK33 pKa = 9.17TRR35 pKa = 11.84KK36 pKa = 8.95PRR38 pKa = 11.84PAPQNNVSWFTPLTQHH54 pKa = 6.68GKK56 pKa = 9.18QALTFNPGQGVPLNANDD73 pKa = 5.2DD74 pKa = 3.75PDD76 pKa = 4.07SRR78 pKa = 11.84IGYY81 pKa = 5.38WRR83 pKa = 11.84RR84 pKa = 11.84NIRR87 pKa = 11.84TVTKK91 pKa = 10.3NGKK94 pKa = 6.92PQKK97 pKa = 10.34LDD99 pKa = 3.39PRR101 pKa = 11.84WYY103 pKa = 10.04FYY105 pKa = 11.53YY106 pKa = 10.65LGTGPEE112 pKa = 3.91QNLKK116 pKa = 10.49YY117 pKa = 10.28LQQKK121 pKa = 10.41DD122 pKa = 3.88GIVWVAAHH130 pKa = 6.71GALNVPVKK138 pKa = 10.02TVGTRR143 pKa = 11.84NPANDD148 pKa = 3.52QAIVAAFAPGTEE160 pKa = 4.22LPKK163 pKa = 10.83GFYY166 pKa = 10.61VEE168 pKa = 5.11GSRR171 pKa = 11.84STSSASSAASSRR183 pKa = 11.84SNSRR187 pKa = 11.84SRR189 pKa = 11.84NSSLSRR195 pKa = 11.84SSSPGRR201 pKa = 11.84GLPPATDD208 pKa = 3.94PNGVLAALLLTKK220 pKa = 10.73LEE222 pKa = 4.29ALDD225 pKa = 4.4AKK227 pKa = 11.27VNGPKK232 pKa = 9.84QPPVVTKK239 pKa = 9.39KK240 pKa = 8.76TAAEE244 pKa = 3.84IAAKK248 pKa = 9.83PRR250 pKa = 11.84QKK252 pKa = 10.25RR253 pKa = 11.84VAHH256 pKa = 6.45KK257 pKa = 10.33GYY259 pKa = 10.37NVNAAYY265 pKa = 10.1GRR267 pKa = 11.84RR268 pKa = 11.84GPGPYY273 pKa = 9.33QGNFGTQEE281 pKa = 3.91FNKK284 pKa = 10.55LGTDD288 pKa = 3.65YY289 pKa = 11.06PKK291 pKa = 10.28WPQIAQLAPTPSAFFGMSRR310 pKa = 11.84FAVQKK315 pKa = 10.75NDD317 pKa = 3.33DD318 pKa = 4.31GTWLTYY324 pKa = 10.12HH325 pKa = 6.3GHH327 pKa = 6.05IKK329 pKa = 9.7MDD331 pKa = 3.35EE332 pKa = 4.13SDD334 pKa = 4.11PNFQVWMTEE343 pKa = 3.81LQQNIDD349 pKa = 3.2AYY351 pKa = 10.32KK352 pKa = 10.5NFPQKK357 pKa = 10.37EE358 pKa = 4.35EE359 pKa = 4.06KK360 pKa = 10.17KK361 pKa = 10.23SRR363 pKa = 11.84KK364 pKa = 9.22PKK366 pKa = 9.07TKK368 pKa = 10.19NVDD371 pKa = 3.33MAPQDD376 pKa = 3.68VMGAAAVDD384 pKa = 4.3LEE386 pKa = 4.49WDD388 pKa = 3.62SSIDD392 pKa = 3.35QTGPNTIVVKK402 pKa = 10.14PKK404 pKa = 10.15KK405 pKa = 8.97QRR407 pKa = 11.84KK408 pKa = 7.3PAADD412 pKa = 3.59NNISEE417 pKa = 4.39II418 pKa = 3.96
MM1 pKa = 7.31AAEE4 pKa = 4.32GQRR7 pKa = 11.84VTFLTQGSNQNNNNQEE23 pKa = 4.12NQDD26 pKa = 3.44GRR28 pKa = 11.84GARR31 pKa = 11.84PKK33 pKa = 9.17TRR35 pKa = 11.84KK36 pKa = 8.95PRR38 pKa = 11.84PAPQNNVSWFTPLTQHH54 pKa = 6.68GKK56 pKa = 9.18QALTFNPGQGVPLNANDD73 pKa = 5.2DD74 pKa = 3.75PDD76 pKa = 4.07SRR78 pKa = 11.84IGYY81 pKa = 5.38WRR83 pKa = 11.84RR84 pKa = 11.84NIRR87 pKa = 11.84TVTKK91 pKa = 10.3NGKK94 pKa = 6.92PQKK97 pKa = 10.34LDD99 pKa = 3.39PRR101 pKa = 11.84WYY103 pKa = 10.04FYY105 pKa = 11.53YY106 pKa = 10.65LGTGPEE112 pKa = 3.91QNLKK116 pKa = 10.49YY117 pKa = 10.28LQQKK121 pKa = 10.41DD122 pKa = 3.88GIVWVAAHH130 pKa = 6.71GALNVPVKK138 pKa = 10.02TVGTRR143 pKa = 11.84NPANDD148 pKa = 3.52QAIVAAFAPGTEE160 pKa = 4.22LPKK163 pKa = 10.83GFYY166 pKa = 10.61VEE168 pKa = 5.11GSRR171 pKa = 11.84STSSASSAASSRR183 pKa = 11.84SNSRR187 pKa = 11.84SRR189 pKa = 11.84NSSLSRR195 pKa = 11.84SSSPGRR201 pKa = 11.84GLPPATDD208 pKa = 3.94PNGVLAALLLTKK220 pKa = 10.73LEE222 pKa = 4.29ALDD225 pKa = 4.4AKK227 pKa = 11.27VNGPKK232 pKa = 9.84QPPVVTKK239 pKa = 9.39KK240 pKa = 8.76TAAEE244 pKa = 3.84IAAKK248 pKa = 9.83PRR250 pKa = 11.84QKK252 pKa = 10.25RR253 pKa = 11.84VAHH256 pKa = 6.45KK257 pKa = 10.33GYY259 pKa = 10.37NVNAAYY265 pKa = 10.1GRR267 pKa = 11.84RR268 pKa = 11.84GPGPYY273 pKa = 9.33QGNFGTQEE281 pKa = 3.91FNKK284 pKa = 10.55LGTDD288 pKa = 3.65YY289 pKa = 11.06PKK291 pKa = 10.28WPQIAQLAPTPSAFFGMSRR310 pKa = 11.84FAVQKK315 pKa = 10.75NDD317 pKa = 3.33DD318 pKa = 4.31GTWLTYY324 pKa = 10.12HH325 pKa = 6.3GHH327 pKa = 6.05IKK329 pKa = 9.7MDD331 pKa = 3.35EE332 pKa = 4.13SDD334 pKa = 4.11PNFQVWMTEE343 pKa = 3.81LQQNIDD349 pKa = 3.2AYY351 pKa = 10.32KK352 pKa = 10.5NFPQKK357 pKa = 10.37EE358 pKa = 4.35EE359 pKa = 4.06KK360 pKa = 10.17KK361 pKa = 10.23SRR363 pKa = 11.84KK364 pKa = 9.22PKK366 pKa = 9.07TKK368 pKa = 10.19NVDD371 pKa = 3.33MAPQDD376 pKa = 3.68VMGAAAVDD384 pKa = 4.3LEE386 pKa = 4.49WDD388 pKa = 3.62SSIDD392 pKa = 3.35QTGPNTIVVKK402 pKa = 10.14PKK404 pKa = 10.15KK405 pKa = 8.97QRR407 pKa = 11.84KK408 pKa = 7.3PAADD412 pKa = 3.59NNISEE417 pKa = 4.39II418 pKa = 3.96
Molecular weight: 45.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10159 |
72 |
7247 |
1128.8 |
126.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.097 ± 0.29 | 3.406 ± 0.444 |
5.375 ± 0.609 | 4.233 ± 0.376 |
4.922 ± 0.318 | 5.571 ± 0.288 |
2.136 ± 0.237 | 4.991 ± 0.409 |
5.512 ± 0.758 | 8.406 ± 0.588 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.49 ± 0.229 | 5.473 ± 0.628 |
4.213 ± 0.606 | 3.642 ± 0.686 |
3.691 ± 0.323 | 6.566 ± 0.298 |
7.668 ± 0.951 | 8.859 ± 0.748 |
1.073 ± 0.165 | 4.676 ± 0.275 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
