
bacterium HR28
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
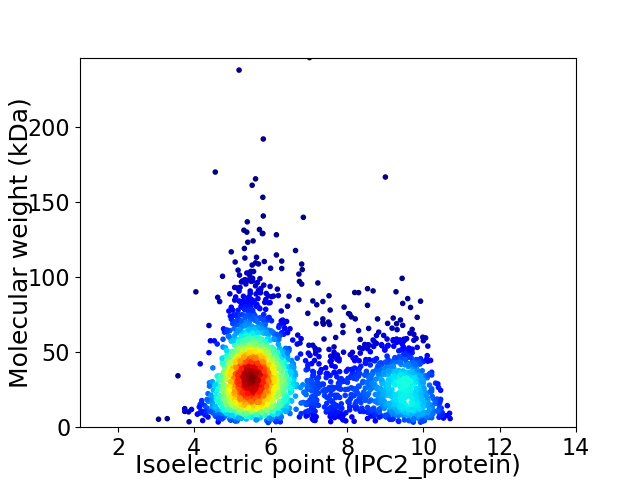
Virtual 2D-PAGE plot for 2802 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5Z228|A0A2H5Z228_9BACT Stage II sporulation protein E OS=bacterium HR28 OX=2035423 GN=spoIIE PE=4 SV=1
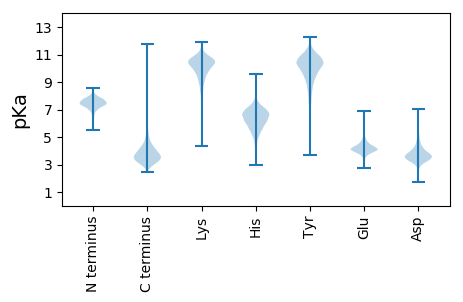
MM1 pKa = 7.65GKK3 pKa = 9.73RR4 pKa = 11.84LVHH7 pKa = 6.62LLLAVAMAVSAFAGLTIAPAGAAQEE32 pKa = 4.54VPSGTIYY39 pKa = 10.53FPWVPNGEE47 pKa = 4.5EE48 pKa = 4.07YY49 pKa = 11.11AGMGPWYY56 pKa = 10.64SSVTIQNFTEE66 pKa = 4.7FPVRR70 pKa = 11.84ANIYY74 pKa = 9.88KK75 pKa = 10.32ADD77 pKa = 3.88GTKK80 pKa = 9.16LTSVTFEE87 pKa = 3.75PWASKK92 pKa = 10.03SWSSADD98 pKa = 3.44LFGNGSGAGVFVDD111 pKa = 4.14AAVGPLTVTRR121 pKa = 11.84GDD123 pKa = 3.33TDD125 pKa = 3.23IDD127 pKa = 4.01SVSVGRR133 pKa = 11.84CVVQSVSISQGATVFVQGVDD153 pKa = 4.07YY154 pKa = 10.57IWSQTGSRR162 pKa = 11.84LDD164 pKa = 3.45ITWIGSAPAAGTDD177 pKa = 3.38YY178 pKa = 10.37TVNVACDD185 pKa = 3.59GLLGGVVKK193 pKa = 10.18MAAPQALISAAWTGTGHH210 pKa = 6.67EE211 pKa = 4.7SVTGYY216 pKa = 9.7TSLPVEE222 pKa = 5.47DD223 pKa = 3.62VWTWPRR229 pKa = 11.84SWVFPIIQTNNGWNSVMHH247 pKa = 6.06ITNFGGGNCSVNVDD261 pKa = 4.19LYY263 pKa = 9.52QTPTRR268 pKa = 11.84AGAPGASGPSEE279 pKa = 3.82GHH281 pKa = 5.87FSEE284 pKa = 5.43LLAAGEE290 pKa = 4.29TWHH293 pKa = 7.12IDD295 pKa = 3.31LVQDD299 pKa = 3.96YY300 pKa = 9.21GWPADD305 pKa = 3.9WIGTAFITADD315 pKa = 3.69CGVAASVDD323 pKa = 3.72RR324 pKa = 11.84LKK326 pKa = 10.9AAQPWGDD333 pKa = 4.01PVNMAVTNVAQPTFGNWSEE352 pKa = 4.41VYY354 pKa = 10.7APLVYY359 pKa = 10.39QNYY362 pKa = 9.64NGWNTGISIVNLDD375 pKa = 3.64PDD377 pKa = 4.31FNNNVQVQFYY387 pKa = 10.3NRR389 pKa = 11.84DD390 pKa = 3.13GTLVHH395 pKa = 6.42SEE397 pKa = 4.3SLTIPPRR404 pKa = 11.84AMEE407 pKa = 4.09WLYY410 pKa = 11.24LPEE413 pKa = 4.23RR414 pKa = 11.84TDD416 pKa = 4.49LGTQGLSQAVIRR428 pKa = 11.84SLSGLNLAAAVDD440 pKa = 4.19SVKK443 pKa = 9.9YY444 pKa = 9.41TGIDD448 pKa = 3.17QDD450 pKa = 3.74VGQATSYY457 pKa = 10.49LAQQGTRR464 pKa = 11.84AARR467 pKa = 11.84PGEE470 pKa = 4.2GAQNGLLSVALFQKK484 pKa = 10.47QFGLTAQQDD493 pKa = 3.84NSGIAYY499 pKa = 9.71FNANTGAPATVEE511 pKa = 3.87VTFYY515 pKa = 10.92DD516 pKa = 3.85GAGFRR521 pKa = 11.84VAPTLVNDD529 pKa = 4.55LVINLPPAGGGILYY543 pKa = 10.01APWYY547 pKa = 10.7GEE549 pKa = 3.89MPFGFRR555 pKa = 11.84GSVVLEE561 pKa = 4.17VTQGGPVACVSNNVNYY577 pKa = 9.74QVQFDD582 pKa = 3.75GTATYY587 pKa = 11.19NCFIRR592 pKa = 11.84EE593 pKa = 4.25AVDD596 pKa = 3.57GAVPRR601 pKa = 11.84PEE603 pKa = 3.76WTLEE607 pKa = 3.72ARR609 pKa = 11.84PANAANLAARR619 pKa = 11.84TVDD622 pKa = 3.13TDD624 pKa = 3.49VEE626 pKa = 4.51RR627 pKa = 11.84FQHH630 pKa = 5.21TVTAVVRR637 pKa = 11.84FAGQPQTGVPVLFEE651 pKa = 4.08ILNGSRR657 pKa = 11.84NDD659 pKa = 4.06DD660 pKa = 3.66PAEE663 pKa = 4.18NDD665 pKa = 3.04QGTYY669 pKa = 10.72NPAGDD674 pKa = 4.46DD675 pKa = 3.4DD676 pKa = 5.03VVVNTNNNGVASWTYY691 pKa = 10.87QDD693 pKa = 3.96GGAANTSGVDD703 pKa = 3.83TILVCADD710 pKa = 3.17VNGNSQCDD718 pKa = 3.5PGEE721 pKa = 3.94PRR723 pKa = 11.84QTVRR727 pKa = 11.84KK728 pKa = 8.17VWATATVDD736 pKa = 3.6QGQPPQDD743 pKa = 3.33AGVVGDD749 pKa = 3.81NTEE752 pKa = 4.23DD753 pKa = 3.26LVLAIDD759 pKa = 4.42VDD761 pKa = 4.3PDD763 pKa = 3.34QLNAGLPLVLRR774 pKa = 11.84IVNEE778 pKa = 4.02TGGDD782 pKa = 3.45VSFAVCNQPDD792 pKa = 3.91RR793 pKa = 11.84NVQPWFVSADD803 pKa = 3.76TNVPLYY809 pKa = 10.3ICADD813 pKa = 3.79PAPAGGTFQVEE824 pKa = 4.7AYY826 pKa = 9.36WDD828 pKa = 3.75VNANGQVDD836 pKa = 3.77AGVDD840 pKa = 3.63VPLIGSPVNFTWW852 pKa = 3.79
MM1 pKa = 7.65GKK3 pKa = 9.73RR4 pKa = 11.84LVHH7 pKa = 6.62LLLAVAMAVSAFAGLTIAPAGAAQEE32 pKa = 4.54VPSGTIYY39 pKa = 10.53FPWVPNGEE47 pKa = 4.5EE48 pKa = 4.07YY49 pKa = 11.11AGMGPWYY56 pKa = 10.64SSVTIQNFTEE66 pKa = 4.7FPVRR70 pKa = 11.84ANIYY74 pKa = 9.88KK75 pKa = 10.32ADD77 pKa = 3.88GTKK80 pKa = 9.16LTSVTFEE87 pKa = 3.75PWASKK92 pKa = 10.03SWSSADD98 pKa = 3.44LFGNGSGAGVFVDD111 pKa = 4.14AAVGPLTVTRR121 pKa = 11.84GDD123 pKa = 3.33TDD125 pKa = 3.23IDD127 pKa = 4.01SVSVGRR133 pKa = 11.84CVVQSVSISQGATVFVQGVDD153 pKa = 4.07YY154 pKa = 10.57IWSQTGSRR162 pKa = 11.84LDD164 pKa = 3.45ITWIGSAPAAGTDD177 pKa = 3.38YY178 pKa = 10.37TVNVACDD185 pKa = 3.59GLLGGVVKK193 pKa = 10.18MAAPQALISAAWTGTGHH210 pKa = 6.67EE211 pKa = 4.7SVTGYY216 pKa = 9.7TSLPVEE222 pKa = 5.47DD223 pKa = 3.62VWTWPRR229 pKa = 11.84SWVFPIIQTNNGWNSVMHH247 pKa = 6.06ITNFGGGNCSVNVDD261 pKa = 4.19LYY263 pKa = 9.52QTPTRR268 pKa = 11.84AGAPGASGPSEE279 pKa = 3.82GHH281 pKa = 5.87FSEE284 pKa = 5.43LLAAGEE290 pKa = 4.29TWHH293 pKa = 7.12IDD295 pKa = 3.31LVQDD299 pKa = 3.96YY300 pKa = 9.21GWPADD305 pKa = 3.9WIGTAFITADD315 pKa = 3.69CGVAASVDD323 pKa = 3.72RR324 pKa = 11.84LKK326 pKa = 10.9AAQPWGDD333 pKa = 4.01PVNMAVTNVAQPTFGNWSEE352 pKa = 4.41VYY354 pKa = 10.7APLVYY359 pKa = 10.39QNYY362 pKa = 9.64NGWNTGISIVNLDD375 pKa = 3.64PDD377 pKa = 4.31FNNNVQVQFYY387 pKa = 10.3NRR389 pKa = 11.84DD390 pKa = 3.13GTLVHH395 pKa = 6.42SEE397 pKa = 4.3SLTIPPRR404 pKa = 11.84AMEE407 pKa = 4.09WLYY410 pKa = 11.24LPEE413 pKa = 4.23RR414 pKa = 11.84TDD416 pKa = 4.49LGTQGLSQAVIRR428 pKa = 11.84SLSGLNLAAAVDD440 pKa = 4.19SVKK443 pKa = 9.9YY444 pKa = 9.41TGIDD448 pKa = 3.17QDD450 pKa = 3.74VGQATSYY457 pKa = 10.49LAQQGTRR464 pKa = 11.84AARR467 pKa = 11.84PGEE470 pKa = 4.2GAQNGLLSVALFQKK484 pKa = 10.47QFGLTAQQDD493 pKa = 3.84NSGIAYY499 pKa = 9.71FNANTGAPATVEE511 pKa = 3.87VTFYY515 pKa = 10.92DD516 pKa = 3.85GAGFRR521 pKa = 11.84VAPTLVNDD529 pKa = 4.55LVINLPPAGGGILYY543 pKa = 10.01APWYY547 pKa = 10.7GEE549 pKa = 3.89MPFGFRR555 pKa = 11.84GSVVLEE561 pKa = 4.17VTQGGPVACVSNNVNYY577 pKa = 9.74QVQFDD582 pKa = 3.75GTATYY587 pKa = 11.19NCFIRR592 pKa = 11.84EE593 pKa = 4.25AVDD596 pKa = 3.57GAVPRR601 pKa = 11.84PEE603 pKa = 3.76WTLEE607 pKa = 3.72ARR609 pKa = 11.84PANAANLAARR619 pKa = 11.84TVDD622 pKa = 3.13TDD624 pKa = 3.49VEE626 pKa = 4.51RR627 pKa = 11.84FQHH630 pKa = 5.21TVTAVVRR637 pKa = 11.84FAGQPQTGVPVLFEE651 pKa = 4.08ILNGSRR657 pKa = 11.84NDD659 pKa = 4.06DD660 pKa = 3.66PAEE663 pKa = 4.18NDD665 pKa = 3.04QGTYY669 pKa = 10.72NPAGDD674 pKa = 4.46DD675 pKa = 3.4DD676 pKa = 5.03VVVNTNNNGVASWTYY691 pKa = 10.87QDD693 pKa = 3.96GGAANTSGVDD703 pKa = 3.83TILVCADD710 pKa = 3.17VNGNSQCDD718 pKa = 3.5PGEE721 pKa = 3.94PRR723 pKa = 11.84QTVRR727 pKa = 11.84KK728 pKa = 8.17VWATATVDD736 pKa = 3.6QGQPPQDD743 pKa = 3.33AGVVGDD749 pKa = 3.81NTEE752 pKa = 4.23DD753 pKa = 3.26LVLAIDD759 pKa = 4.42VDD761 pKa = 4.3PDD763 pKa = 3.34QLNAGLPLVLRR774 pKa = 11.84IVNEE778 pKa = 4.02TGGDD782 pKa = 3.45VSFAVCNQPDD792 pKa = 3.91RR793 pKa = 11.84NVQPWFVSADD803 pKa = 3.76TNVPLYY809 pKa = 10.3ICADD813 pKa = 3.79PAPAGGTFQVEE824 pKa = 4.7AYY826 pKa = 9.36WDD828 pKa = 3.75VNANGQVDD836 pKa = 3.77AGVDD840 pKa = 3.63VPLIGSPVNFTWW852 pKa = 3.79
Molecular weight: 90.26 kDa
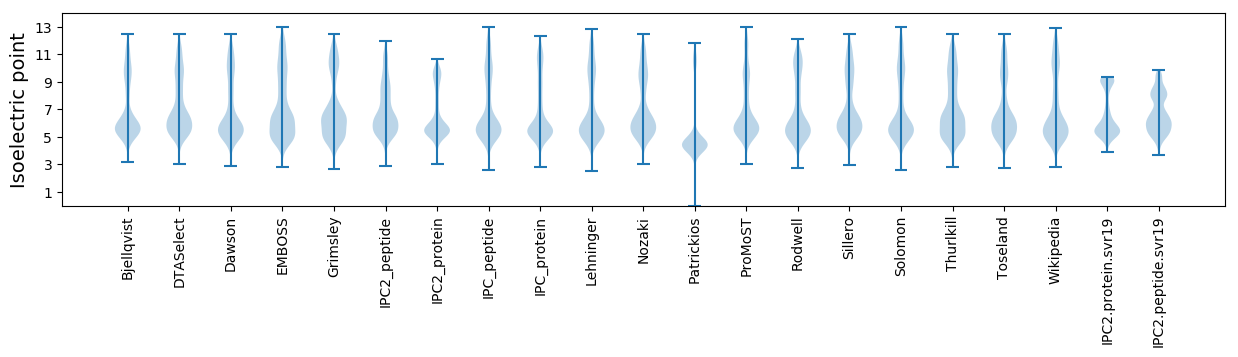
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5Z8G3|A0A2H5Z8G3_9BACT Cyclic nucleotide-binding domain-containing protein OS=bacterium HR28 OX=2035423 GN=HRbin28_02463 PE=4 SV=1
MM1 pKa = 7.51AKK3 pKa = 9.86KK4 pKa = 10.39AKK6 pKa = 9.54ADD8 pKa = 4.49RR9 pKa = 11.84IIITLEE15 pKa = 3.78CTVCRR20 pKa = 11.84EE21 pKa = 4.05RR22 pKa = 11.84NYY24 pKa = 9.2VTQKK28 pKa = 10.25NRR30 pKa = 11.84RR31 pKa = 11.84NDD33 pKa = 3.56PGRR36 pKa = 11.84LEE38 pKa = 3.88LRR40 pKa = 11.84KK41 pKa = 9.79YY42 pKa = 10.31CPRR45 pKa = 11.84CRR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 4.32QVHH52 pKa = 6.67RR53 pKa = 11.84EE54 pKa = 3.63TRR56 pKa = 3.29
MM1 pKa = 7.51AKK3 pKa = 9.86KK4 pKa = 10.39AKK6 pKa = 9.54ADD8 pKa = 4.49RR9 pKa = 11.84IIITLEE15 pKa = 3.78CTVCRR20 pKa = 11.84EE21 pKa = 4.05RR22 pKa = 11.84NYY24 pKa = 9.2VTQKK28 pKa = 10.25NRR30 pKa = 11.84RR31 pKa = 11.84NDD33 pKa = 3.56PGRR36 pKa = 11.84LEE38 pKa = 3.88LRR40 pKa = 11.84KK41 pKa = 9.79YY42 pKa = 10.31CPRR45 pKa = 11.84CRR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 4.32QVHH52 pKa = 6.67RR53 pKa = 11.84EE54 pKa = 3.63TRR56 pKa = 3.29
Molecular weight: 6.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
898898 |
29 |
2214 |
320.8 |
35.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.497 ± 0.056 | 0.822 ± 0.016 |
4.607 ± 0.026 | 6.764 ± 0.052 |
3.267 ± 0.027 | 7.986 ± 0.048 |
2.074 ± 0.021 | 4.624 ± 0.032 |
1.624 ± 0.026 | 11.854 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.646 ± 0.017 | 1.81 ± 0.02 |
6.156 ± 0.038 | 3.578 ± 0.031 |
8.931 ± 0.048 | 4.541 ± 0.035 |
5.469 ± 0.036 | 8.641 ± 0.045 |
1.726 ± 0.023 | 2.383 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
