
Bosea caraganae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Boseaceae; Bosea
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
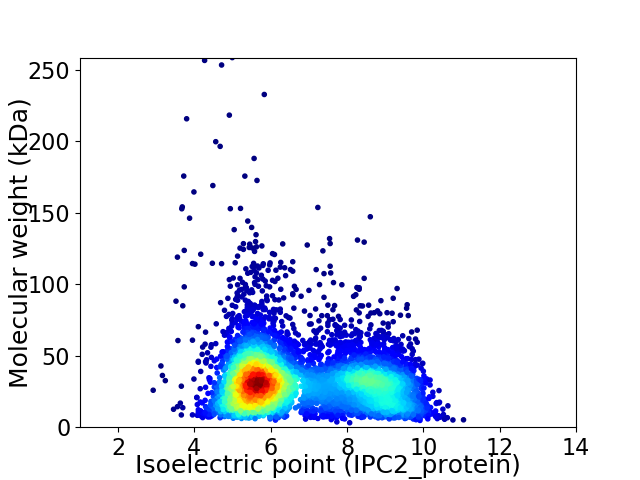
Virtual 2D-PAGE plot for 5639 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A370L739|A0A370L739_9BRAD Uncharacterized protein OS=Bosea caraganae OX=2763117 GN=DWE98_11885 PE=4 SV=1
MM1 pKa = 7.22TGAAGGAGGGEE12 pKa = 4.16NASGSAGGSGSGTAGGIGGPGGGSVGGTGGNGGGGATGGAGTADD56 pKa = 3.48SGSNGGLGGGGGGGAALVLNGSSLVVTTSGGVTGGIGGAGGGANSSFSGNLGNGGGGGAGIASASTGLQVSNSGTITGGAGGDD139 pKa = 3.2AGFLGFLGSGATNGVGGAGGTGVSLSGASASFTNTGAVRR178 pKa = 11.84GGGGGLLNTLNSFTGSGAGGDD199 pKa = 3.69GGAGLVISGAGGTVSNSGTIGGGTGGYY226 pKa = 10.45DD227 pKa = 3.12IDD229 pKa = 4.07WNVSQGGASGAGVILGGSGASLATGTVLTNNAGATIVGANRR270 pKa = 11.84AVMLAASGPDD280 pKa = 3.15NGGAGVSITGNYY292 pKa = 7.19NTINNAGTITGGRR305 pKa = 11.84AGNAGTAGVLVSGDD319 pKa = 3.37NNLVVSSGTITGGGAGGTIAPSVQFDD345 pKa = 3.42GHH347 pKa = 6.45NNTLEE352 pKa = 3.88LRR354 pKa = 11.84AGYY357 pKa = 10.65ALGGGAKK364 pKa = 10.36VNGTGGTLVLGGTANAAFDD383 pKa = 3.97LARR386 pKa = 11.84IGTSATSFNGFSAFNKK402 pKa = 9.39TGGSTWTLNGIYY414 pKa = 9.88TDD416 pKa = 3.51TSNWNVQQGTLAMASLAEE434 pKa = 4.25TTGTVSVASGAALSVLALSRR454 pKa = 11.84VHH456 pKa = 6.08GQVTIANGGTLVTTTTTSAAALDD479 pKa = 4.17GLTLSPNSILEE490 pKa = 4.21MNVTRR495 pKa = 11.84AGNTTGLLTLGSGGLVLDD513 pKa = 3.96GTLNITDD520 pKa = 4.16SGNALAAGTYY530 pKa = 10.41RR531 pKa = 11.84LFNVIGTLTNNTMVIGPAPMQYY553 pKa = 10.32LYY555 pKa = 10.98SIDD558 pKa = 3.3TSNPVFITLLVASNTSYY575 pKa = 11.09WNGAQATPNGTVNGGSGTWDD595 pKa = 2.94AAATNWTNATGTTSKK610 pKa = 10.86AWGGYY615 pKa = 6.98EE616 pKa = 4.69AVFQGTPGTVTVDD629 pKa = 3.11NGGVSAYY636 pKa = 8.02LTHH639 pKa = 6.77FLVDD643 pKa = 4.7GYY645 pKa = 8.73TLSGGPLTLASLSAAKK661 pKa = 10.02PIVEE665 pKa = 4.37VATGATATIQSVIAGTAGLDD685 pKa = 3.29KK686 pKa = 10.8TGAGTLTLTGANTYY700 pKa = 8.95TGNTSITAGTLQIAGSGSILGDD722 pKa = 3.39VANSGTLSFNPGSDD736 pKa = 2.63RR737 pKa = 11.84AFAGNISGTGSLTKK751 pKa = 10.65LGGSTLTLSIANTYY765 pKa = 10.77AGGTTVSAGVLNVRR779 pKa = 11.84NDD781 pKa = 3.31SALGTGAVSVASGAALEE798 pKa = 4.24VQGYY802 pKa = 10.12ISLPNALTLSGTGVANGGALRR823 pKa = 11.84NVSGANGLDD832 pKa = 3.69GAITLAGDD840 pKa = 3.38TRR842 pKa = 11.84INNDD846 pKa = 3.51SFDD849 pKa = 3.99SDD851 pKa = 3.68LGLVGGISGSGHH863 pKa = 5.01TLVLGGSSEE872 pKa = 4.81YY873 pKa = 9.0ITSVTGAIATGLGGVTIDD891 pKa = 3.29VTGAAGRR898 pKa = 11.84VILAGANTYY907 pKa = 9.92TGVTTVNSGTLQLMYY922 pKa = 10.53GAAIADD928 pKa = 3.76TGAVVVNAPGSLQIYY943 pKa = 9.14NSEE946 pKa = 4.6TIGSLAGAGSVVLNSGSTLAVGGDD970 pKa = 3.79DD971 pKa = 4.21SSTSFSGVISQSSGVGSLTKK991 pKa = 10.2IGTGSFTLSGTNTYY1005 pKa = 9.58TGATQVDD1012 pKa = 4.1GGEE1015 pKa = 4.03LRR1017 pKa = 11.84VNGSVQGTVGVASGATLGGSGAIAGAVTVASGGTLSAGSSPGTLTVGSLEE1067 pKa = 4.44LNSGANTVFEE1077 pKa = 4.88LNSPGVVGGGTNDD1090 pKa = 3.86HH1091 pKa = 6.95IIVNGSGAAGNLVLGGTLTANVASAGYY1118 pKa = 9.9YY1119 pKa = 10.26RR1120 pKa = 11.84LFDD1123 pKa = 3.6VTGGGAISGSFDD1135 pKa = 3.4TLALTAPSVAGATGTVYY1152 pKa = 10.58NAPSGVPTQVNLAVLGTSQAMQFWDD1177 pKa = 4.65GSDD1180 pKa = 3.2QLGNGTVDD1188 pKa = 3.6GGAGTWNAGNANWTGAPGQADD1209 pKa = 4.89FNAPWSDD1216 pKa = 3.41SVGVFF1221 pKa = 3.78
MM1 pKa = 7.22TGAAGGAGGGEE12 pKa = 4.16NASGSAGGSGSGTAGGIGGPGGGSVGGTGGNGGGGATGGAGTADD56 pKa = 3.48SGSNGGLGGGGGGGAALVLNGSSLVVTTSGGVTGGIGGAGGGANSSFSGNLGNGGGGGAGIASASTGLQVSNSGTITGGAGGDD139 pKa = 3.2AGFLGFLGSGATNGVGGAGGTGVSLSGASASFTNTGAVRR178 pKa = 11.84GGGGGLLNTLNSFTGSGAGGDD199 pKa = 3.69GGAGLVISGAGGTVSNSGTIGGGTGGYY226 pKa = 10.45DD227 pKa = 3.12IDD229 pKa = 4.07WNVSQGGASGAGVILGGSGASLATGTVLTNNAGATIVGANRR270 pKa = 11.84AVMLAASGPDD280 pKa = 3.15NGGAGVSITGNYY292 pKa = 7.19NTINNAGTITGGRR305 pKa = 11.84AGNAGTAGVLVSGDD319 pKa = 3.37NNLVVSSGTITGGGAGGTIAPSVQFDD345 pKa = 3.42GHH347 pKa = 6.45NNTLEE352 pKa = 3.88LRR354 pKa = 11.84AGYY357 pKa = 10.65ALGGGAKK364 pKa = 10.36VNGTGGTLVLGGTANAAFDD383 pKa = 3.97LARR386 pKa = 11.84IGTSATSFNGFSAFNKK402 pKa = 9.39TGGSTWTLNGIYY414 pKa = 9.88TDD416 pKa = 3.51TSNWNVQQGTLAMASLAEE434 pKa = 4.25TTGTVSVASGAALSVLALSRR454 pKa = 11.84VHH456 pKa = 6.08GQVTIANGGTLVTTTTTSAAALDD479 pKa = 4.17GLTLSPNSILEE490 pKa = 4.21MNVTRR495 pKa = 11.84AGNTTGLLTLGSGGLVLDD513 pKa = 3.96GTLNITDD520 pKa = 4.16SGNALAAGTYY530 pKa = 10.41RR531 pKa = 11.84LFNVIGTLTNNTMVIGPAPMQYY553 pKa = 10.32LYY555 pKa = 10.98SIDD558 pKa = 3.3TSNPVFITLLVASNTSYY575 pKa = 11.09WNGAQATPNGTVNGGSGTWDD595 pKa = 2.94AAATNWTNATGTTSKK610 pKa = 10.86AWGGYY615 pKa = 6.98EE616 pKa = 4.69AVFQGTPGTVTVDD629 pKa = 3.11NGGVSAYY636 pKa = 8.02LTHH639 pKa = 6.77FLVDD643 pKa = 4.7GYY645 pKa = 8.73TLSGGPLTLASLSAAKK661 pKa = 10.02PIVEE665 pKa = 4.37VATGATATIQSVIAGTAGLDD685 pKa = 3.29KK686 pKa = 10.8TGAGTLTLTGANTYY700 pKa = 8.95TGNTSITAGTLQIAGSGSILGDD722 pKa = 3.39VANSGTLSFNPGSDD736 pKa = 2.63RR737 pKa = 11.84AFAGNISGTGSLTKK751 pKa = 10.65LGGSTLTLSIANTYY765 pKa = 10.77AGGTTVSAGVLNVRR779 pKa = 11.84NDD781 pKa = 3.31SALGTGAVSVASGAALEE798 pKa = 4.24VQGYY802 pKa = 10.12ISLPNALTLSGTGVANGGALRR823 pKa = 11.84NVSGANGLDD832 pKa = 3.69GAITLAGDD840 pKa = 3.38TRR842 pKa = 11.84INNDD846 pKa = 3.51SFDD849 pKa = 3.99SDD851 pKa = 3.68LGLVGGISGSGHH863 pKa = 5.01TLVLGGSSEE872 pKa = 4.81YY873 pKa = 9.0ITSVTGAIATGLGGVTIDD891 pKa = 3.29VTGAAGRR898 pKa = 11.84VILAGANTYY907 pKa = 9.92TGVTTVNSGTLQLMYY922 pKa = 10.53GAAIADD928 pKa = 3.76TGAVVVNAPGSLQIYY943 pKa = 9.14NSEE946 pKa = 4.6TIGSLAGAGSVVLNSGSTLAVGGDD970 pKa = 3.79DD971 pKa = 4.21SSTSFSGVISQSSGVGSLTKK991 pKa = 10.2IGTGSFTLSGTNTYY1005 pKa = 9.58TGATQVDD1012 pKa = 4.1GGEE1015 pKa = 4.03LRR1017 pKa = 11.84VNGSVQGTVGVASGATLGGSGAIAGAVTVASGGTLSAGSSPGTLTVGSLEE1067 pKa = 4.44LNSGANTVFEE1077 pKa = 4.88LNSPGVVGGGTNDD1090 pKa = 3.86HH1091 pKa = 6.95IIVNGSGAAGNLVLGGTLTANVASAGYY1118 pKa = 9.9YY1119 pKa = 10.26RR1120 pKa = 11.84LFDD1123 pKa = 3.6VTGGGAISGSFDD1135 pKa = 3.4TLALTAPSVAGATGTVYY1152 pKa = 10.58NAPSGVPTQVNLAVLGTSQAMQFWDD1177 pKa = 4.65GSDD1180 pKa = 3.2QLGNGTVDD1188 pKa = 3.6GGAGTWNAGNANWTGAPGQADD1209 pKa = 4.89FNAPWSDD1216 pKa = 3.41SVGVFF1221 pKa = 3.78
Molecular weight: 113.95 kDa
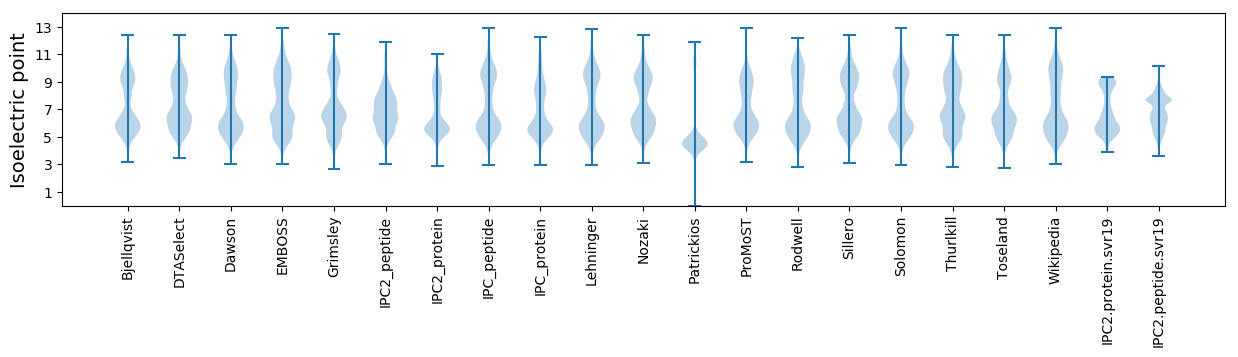
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A370L5N6|A0A370L5N6_9BRAD L-glyceraldehyde 3-phosphate reductase OS=Bosea caraganae OX=2763117 GN=DWE98_12800 PE=4 SV=1
MM1 pKa = 7.88LGQTKK6 pKa = 9.07EE7 pKa = 3.54WWLEE11 pKa = 3.71PSIPPIWFRR20 pKa = 11.84FIMRR24 pKa = 11.84QFALAALLAVCCPAFSFANVPLEE47 pKa = 3.91QVSFQPQVRR56 pKa = 11.84GPGCLKK62 pKa = 10.45PEE64 pKa = 3.99AVAMIRR70 pKa = 11.84EE71 pKa = 4.76LVAKK75 pKa = 10.02IGPIQITSTCGGRR88 pKa = 11.84HH89 pKa = 5.23ARR91 pKa = 11.84RR92 pKa = 11.84SQHH95 pKa = 5.2YY96 pKa = 9.6SGNAVDD102 pKa = 4.9FRR104 pKa = 11.84PMATSARR111 pKa = 11.84KK112 pKa = 9.11AVAVAKK118 pKa = 10.35ALEE121 pKa = 4.35STGGVGSYY129 pKa = 10.65PNGVVHH135 pKa = 6.75VDD137 pKa = 2.98VGDD140 pKa = 4.53RR141 pKa = 11.84EE142 pKa = 4.38ISWFGHH148 pKa = 4.14KK149 pKa = 9.93RR150 pKa = 11.84ARR152 pKa = 11.84TRR154 pKa = 11.84LAYY157 pKa = 10.05AGRR160 pKa = 11.84SRR162 pKa = 11.84RR163 pKa = 3.65
MM1 pKa = 7.88LGQTKK6 pKa = 9.07EE7 pKa = 3.54WWLEE11 pKa = 3.71PSIPPIWFRR20 pKa = 11.84FIMRR24 pKa = 11.84QFALAALLAVCCPAFSFANVPLEE47 pKa = 3.91QVSFQPQVRR56 pKa = 11.84GPGCLKK62 pKa = 10.45PEE64 pKa = 3.99AVAMIRR70 pKa = 11.84EE71 pKa = 4.76LVAKK75 pKa = 10.02IGPIQITSTCGGRR88 pKa = 11.84HH89 pKa = 5.23ARR91 pKa = 11.84RR92 pKa = 11.84SQHH95 pKa = 5.2YY96 pKa = 9.6SGNAVDD102 pKa = 4.9FRR104 pKa = 11.84PMATSARR111 pKa = 11.84KK112 pKa = 9.11AVAVAKK118 pKa = 10.35ALEE121 pKa = 4.35STGGVGSYY129 pKa = 10.65PNGVVHH135 pKa = 6.75VDD137 pKa = 2.98VGDD140 pKa = 4.53RR141 pKa = 11.84EE142 pKa = 4.38ISWFGHH148 pKa = 4.14KK149 pKa = 9.93RR150 pKa = 11.84ARR152 pKa = 11.84TRR154 pKa = 11.84LAYY157 pKa = 10.05AGRR160 pKa = 11.84SRR162 pKa = 11.84RR163 pKa = 3.65
Molecular weight: 17.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1791978 |
28 |
2763 |
317.8 |
34.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.26 ± 0.055 | 0.76 ± 0.01 |
5.332 ± 0.026 | 5.516 ± 0.033 |
3.703 ± 0.022 | 8.929 ± 0.054 |
1.875 ± 0.017 | 5.202 ± 0.023 |
3.236 ± 0.029 | 10.522 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.329 ± 0.016 | 2.446 ± 0.021 |
5.336 ± 0.025 | 3.088 ± 0.018 |
7.071 ± 0.039 | 5.419 ± 0.026 |
5.201 ± 0.032 | 7.374 ± 0.027 |
1.291 ± 0.012 | 2.111 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
