
Winogradskyella endarachnes
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Winogradskyella
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
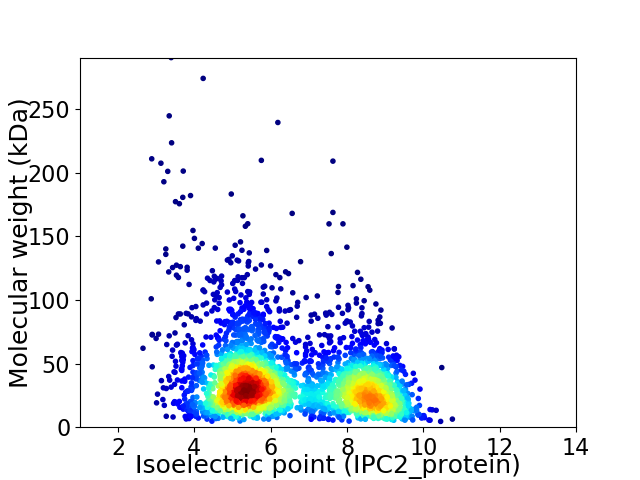
Virtual 2D-PAGE plot for 3122 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6L6U7W8|A0A6L6U7W8_9FLAO Thioredoxin domain-containing protein OS=Winogradskyella endarachnes OX=2681965 GN=GN138_00860 PE=4 SV=1
MM1 pKa = 7.36TKK3 pKa = 10.23YY4 pKa = 10.58FKK6 pKa = 10.94LFLFLFVVTITSVTVVSCAEE26 pKa = 4.27DD27 pKa = 3.73GVNGLDD33 pKa = 3.93GVDD36 pKa = 4.83GIDD39 pKa = 4.79GEE41 pKa = 4.56DD42 pKa = 4.08GEE44 pKa = 5.47DD45 pKa = 4.65GEE47 pKa = 5.57DD48 pKa = 3.45LTIEE52 pKa = 3.87PTIFEE57 pKa = 4.79NKK59 pKa = 9.84SSLQPLVSIQPQFNYY74 pKa = 10.16VEE76 pKa = 4.58AYY78 pKa = 10.5SLISSTDD85 pKa = 3.17EE86 pKa = 3.9LADD89 pKa = 3.74GFKK92 pKa = 11.08LLGAQDD98 pKa = 3.85GAGFLQDD105 pKa = 3.26GDD107 pKa = 3.94GYY109 pKa = 10.35IYY111 pKa = 10.84VVNSEE116 pKa = 4.47DD117 pKa = 3.3DD118 pKa = 3.64HH119 pKa = 7.61AVSRR123 pKa = 11.84IHH125 pKa = 6.85FDD127 pKa = 2.85EE128 pKa = 4.27YY129 pKa = 10.9MNPTGGEE136 pKa = 3.83WLLTDD141 pKa = 4.89DD142 pKa = 4.41VADD145 pKa = 4.15EE146 pKa = 4.55ARR148 pKa = 11.84QCSATMWEE156 pKa = 4.18AAIHH160 pKa = 6.3GGSQDD165 pKa = 3.05IFLSASEE172 pKa = 4.46SINYY176 pKa = 8.62DD177 pKa = 3.25VKK179 pKa = 11.41GIDD182 pKa = 3.62PWVEE186 pKa = 4.2TPTPTADD193 pKa = 3.7FGLDD197 pKa = 3.22ALGEE201 pKa = 4.24FSWEE205 pKa = 4.04NAVPLPKK212 pKa = 9.94DD213 pKa = 3.3AYY215 pKa = 10.24SGKK218 pKa = 8.28TVIIGGDD225 pKa = 3.61DD226 pKa = 3.63DD227 pKa = 5.48SSGSEE232 pKa = 4.05GQVILYY238 pKa = 10.09YY239 pKa = 10.42SEE241 pKa = 5.72NGDD244 pKa = 3.78ADD246 pKa = 3.86LTGGKK251 pKa = 9.38IYY253 pKa = 10.49VLKK256 pKa = 10.69LKK258 pKa = 10.13EE259 pKa = 4.07VSNSAGGVQDD269 pKa = 3.74VVAGTIYY276 pKa = 10.94NEE278 pKa = 4.36SNLDD282 pKa = 3.53LQVSYY287 pKa = 10.56EE288 pKa = 4.08VEE290 pKa = 3.86FVEE293 pKa = 4.79IVNGAAMTKK302 pKa = 10.59NEE304 pKa = 4.25MEE306 pKa = 4.49DD307 pKa = 3.12ACVAVQASAFMRR319 pKa = 11.84VEE321 pKa = 3.89DD322 pKa = 3.72VDD324 pKa = 3.94YY325 pKa = 11.6QKK327 pKa = 11.71GDD329 pKa = 3.53DD330 pKa = 3.62ANARR334 pKa = 11.84NVFFAVTGRR343 pKa = 11.84GPNANTYY350 pKa = 9.97NDD352 pKa = 2.99WGTIYY357 pKa = 10.93KK358 pKa = 10.5LEE360 pKa = 5.23LDD362 pKa = 4.69DD363 pKa = 6.63DD364 pKa = 4.57SPLTGSLTQIVSGNTEE380 pKa = 3.92TNNADD385 pKa = 3.64GNNSLLQSPDD395 pKa = 3.75NICVTEE401 pKa = 4.0NFVYY405 pKa = 10.65YY406 pKa = 10.52QEE408 pKa = 5.3DD409 pKa = 3.86PNSFDD414 pKa = 4.18RR415 pKa = 11.84NHH417 pKa = 6.81AAYY420 pKa = 9.4IYY422 pKa = 7.53QTNLNGDD429 pKa = 3.55NSHH432 pKa = 6.4VVLEE436 pKa = 4.1LLVRR440 pKa = 11.84QDD442 pKa = 4.28LDD444 pKa = 3.73PTGSTGYY451 pKa = 10.06SGEE454 pKa = 4.08FGALIDD460 pKa = 4.18VSDD463 pKa = 4.97KK464 pKa = 11.15LGQPDD469 pKa = 4.11TFMLALQPHH478 pKa = 5.67YY479 pKa = 10.04WEE481 pKa = 5.1SDD483 pKa = 3.41DD484 pKa = 4.96FVVDD488 pKa = 5.05GLPDD492 pKa = 3.77DD493 pKa = 3.73QGGQIILLKK502 pKa = 10.5GLPRR506 pKa = 5.12
MM1 pKa = 7.36TKK3 pKa = 10.23YY4 pKa = 10.58FKK6 pKa = 10.94LFLFLFVVTITSVTVVSCAEE26 pKa = 4.27DD27 pKa = 3.73GVNGLDD33 pKa = 3.93GVDD36 pKa = 4.83GIDD39 pKa = 4.79GEE41 pKa = 4.56DD42 pKa = 4.08GEE44 pKa = 5.47DD45 pKa = 4.65GEE47 pKa = 5.57DD48 pKa = 3.45LTIEE52 pKa = 3.87PTIFEE57 pKa = 4.79NKK59 pKa = 9.84SSLQPLVSIQPQFNYY74 pKa = 10.16VEE76 pKa = 4.58AYY78 pKa = 10.5SLISSTDD85 pKa = 3.17EE86 pKa = 3.9LADD89 pKa = 3.74GFKK92 pKa = 11.08LLGAQDD98 pKa = 3.85GAGFLQDD105 pKa = 3.26GDD107 pKa = 3.94GYY109 pKa = 10.35IYY111 pKa = 10.84VVNSEE116 pKa = 4.47DD117 pKa = 3.3DD118 pKa = 3.64HH119 pKa = 7.61AVSRR123 pKa = 11.84IHH125 pKa = 6.85FDD127 pKa = 2.85EE128 pKa = 4.27YY129 pKa = 10.9MNPTGGEE136 pKa = 3.83WLLTDD141 pKa = 4.89DD142 pKa = 4.41VADD145 pKa = 4.15EE146 pKa = 4.55ARR148 pKa = 11.84QCSATMWEE156 pKa = 4.18AAIHH160 pKa = 6.3GGSQDD165 pKa = 3.05IFLSASEE172 pKa = 4.46SINYY176 pKa = 8.62DD177 pKa = 3.25VKK179 pKa = 11.41GIDD182 pKa = 3.62PWVEE186 pKa = 4.2TPTPTADD193 pKa = 3.7FGLDD197 pKa = 3.22ALGEE201 pKa = 4.24FSWEE205 pKa = 4.04NAVPLPKK212 pKa = 9.94DD213 pKa = 3.3AYY215 pKa = 10.24SGKK218 pKa = 8.28TVIIGGDD225 pKa = 3.61DD226 pKa = 3.63DD227 pKa = 5.48SSGSEE232 pKa = 4.05GQVILYY238 pKa = 10.09YY239 pKa = 10.42SEE241 pKa = 5.72NGDD244 pKa = 3.78ADD246 pKa = 3.86LTGGKK251 pKa = 9.38IYY253 pKa = 10.49VLKK256 pKa = 10.69LKK258 pKa = 10.13EE259 pKa = 4.07VSNSAGGVQDD269 pKa = 3.74VVAGTIYY276 pKa = 10.94NEE278 pKa = 4.36SNLDD282 pKa = 3.53LQVSYY287 pKa = 10.56EE288 pKa = 4.08VEE290 pKa = 3.86FVEE293 pKa = 4.79IVNGAAMTKK302 pKa = 10.59NEE304 pKa = 4.25MEE306 pKa = 4.49DD307 pKa = 3.12ACVAVQASAFMRR319 pKa = 11.84VEE321 pKa = 3.89DD322 pKa = 3.72VDD324 pKa = 3.94YY325 pKa = 11.6QKK327 pKa = 11.71GDD329 pKa = 3.53DD330 pKa = 3.62ANARR334 pKa = 11.84NVFFAVTGRR343 pKa = 11.84GPNANTYY350 pKa = 9.97NDD352 pKa = 2.99WGTIYY357 pKa = 10.93KK358 pKa = 10.5LEE360 pKa = 5.23LDD362 pKa = 4.69DD363 pKa = 6.63DD364 pKa = 4.57SPLTGSLTQIVSGNTEE380 pKa = 3.92TNNADD385 pKa = 3.64GNNSLLQSPDD395 pKa = 3.75NICVTEE401 pKa = 4.0NFVYY405 pKa = 10.65YY406 pKa = 10.52QEE408 pKa = 5.3DD409 pKa = 3.86PNSFDD414 pKa = 4.18RR415 pKa = 11.84NHH417 pKa = 6.81AAYY420 pKa = 9.4IYY422 pKa = 7.53QTNLNGDD429 pKa = 3.55NSHH432 pKa = 6.4VVLEE436 pKa = 4.1LLVRR440 pKa = 11.84QDD442 pKa = 4.28LDD444 pKa = 3.73PTGSTGYY451 pKa = 10.06SGEE454 pKa = 4.08FGALIDD460 pKa = 4.18VSDD463 pKa = 4.97KK464 pKa = 11.15LGQPDD469 pKa = 4.11TFMLALQPHH478 pKa = 5.67YY479 pKa = 10.04WEE481 pKa = 5.1SDD483 pKa = 3.41DD484 pKa = 4.96FVVDD488 pKa = 5.05GLPDD492 pKa = 3.77DD493 pKa = 3.73QGGQIILLKK502 pKa = 10.5GLPRR506 pKa = 5.12
Molecular weight: 54.95 kDa
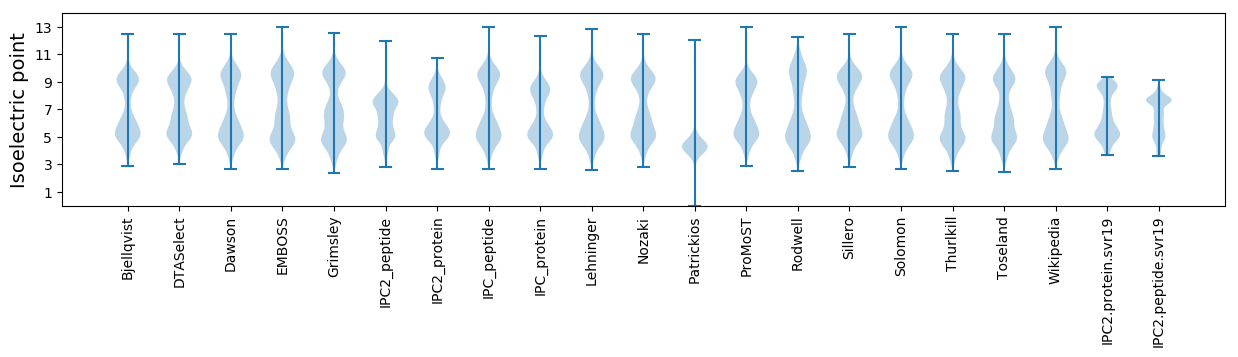
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6L6UBG5|A0A6L6UBG5_9FLAO Uncharacterized protein OS=Winogradskyella endarachnes OX=2681965 GN=GN138_12760 PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.65LSVSTEE48 pKa = 3.95SRR50 pKa = 11.84HH51 pKa = 6.01KK52 pKa = 10.6KK53 pKa = 9.6
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.65LSVSTEE48 pKa = 3.95SRR50 pKa = 11.84HH51 pKa = 6.01KK52 pKa = 10.6KK53 pKa = 9.6
Molecular weight: 6.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1080222 |
38 |
2737 |
346.0 |
39.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.25 ± 0.04 | 0.753 ± 0.014 |
5.833 ± 0.035 | 6.595 ± 0.045 |
5.196 ± 0.036 | 6.185 ± 0.049 |
1.694 ± 0.021 | 8.222 ± 0.04 |
7.74 ± 0.067 | 9.243 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.054 ± 0.021 | 6.585 ± 0.048 |
3.171 ± 0.022 | 3.231 ± 0.019 |
3.105 ± 0.035 | 6.687 ± 0.042 |
6.156 ± 0.059 | 6.183 ± 0.033 |
0.963 ± 0.015 | 4.156 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
