
Bovine faeces associated circular DNA virus 1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
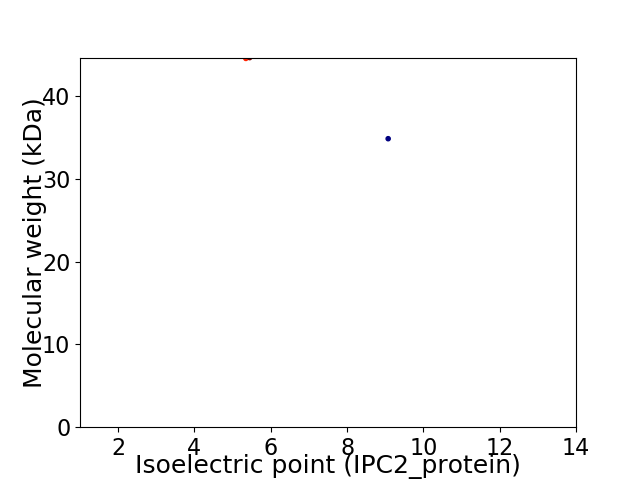
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A160HWJ0|A0A160HWJ0_9VIRU Putative capsid protein OS=Bovine faeces associated circular DNA virus 1 OX=1843764 PE=4 SV=1
MM1 pKa = 8.05SDD3 pKa = 2.89RR4 pKa = 11.84TKK6 pKa = 11.08AKK8 pKa = 9.76RR9 pKa = 11.84WCFTINNPFNVDD21 pKa = 4.32PIIASPGAEE30 pKa = 4.7DD31 pKa = 5.73DD32 pKa = 3.69PDD34 pKa = 3.82EE35 pKa = 4.59PVVIDD40 pKa = 3.52LRR42 pKa = 11.84YY43 pKa = 9.13PFWYY47 pKa = 8.75HH48 pKa = 5.81HH49 pKa = 6.85AMLAVFKK56 pKa = 10.62VEE58 pKa = 3.94YY59 pKa = 9.55MVAQIEE65 pKa = 4.3KK66 pKa = 10.02GEE68 pKa = 4.24NGTPHH73 pKa = 5.85WQGFIIFKK81 pKa = 9.36DD82 pKa = 3.62QKK84 pKa = 10.13RR85 pKa = 11.84LQWLKK90 pKa = 11.02DD91 pKa = 3.52HH92 pKa = 7.0LDD94 pKa = 3.69KK95 pKa = 10.86RR96 pKa = 11.84AHH98 pKa = 5.89WEE100 pKa = 3.89VARR103 pKa = 11.84GTNQQASEE111 pKa = 4.33YY112 pKa = 8.85CKK114 pKa = 10.4KK115 pKa = 10.84DD116 pKa = 3.19EE117 pKa = 4.62TYY119 pKa = 10.83DD120 pKa = 3.64PSVIPADD127 pKa = 3.4MKK129 pKa = 10.35IRR131 pKa = 11.84GRR133 pKa = 11.84FEE135 pKa = 3.76WGSMPEE141 pKa = 3.88RR142 pKa = 11.84NVPKK146 pKa = 10.14KK147 pKa = 10.38DD148 pKa = 3.41EE149 pKa = 4.21RR150 pKa = 11.84LKK152 pKa = 11.1LAADD156 pKa = 3.79EE157 pKa = 5.07LDD159 pKa = 4.08TIKK162 pKa = 10.96DD163 pKa = 3.72GYY165 pKa = 10.09KK166 pKa = 9.89RR167 pKa = 11.84PAEE170 pKa = 4.18IDD172 pKa = 3.56SLTLMQCGFIPAYY185 pKa = 9.82KK186 pKa = 10.23EE187 pKa = 3.55LTADD191 pKa = 2.64ILGPYY196 pKa = 9.07RR197 pKa = 11.84PKK199 pKa = 10.82LQIITMIGPPGTGKK213 pKa = 10.4SYY215 pKa = 10.78AIQQYY220 pKa = 8.88FPNHH224 pKa = 5.6GRR226 pKa = 11.84CICGNNGVWFQNPTAPVMVFEE247 pKa = 4.71EE248 pKa = 4.71FCGQIQLQRR257 pKa = 11.84MLQYY261 pKa = 11.0LDD263 pKa = 4.6PYY265 pKa = 10.57PLALEE270 pKa = 4.65VKK272 pKa = 9.95GGMRR276 pKa = 11.84PAMYY280 pKa = 7.83EE281 pKa = 3.97TVIITSNTRR290 pKa = 11.84PDD292 pKa = 2.63GWYY295 pKa = 9.09TGDD298 pKa = 3.87QAGQPGKK305 pKa = 9.5RR306 pKa = 11.84TDD308 pKa = 3.44ALLALWDD315 pKa = 3.81RR316 pKa = 11.84LGFEE320 pKa = 4.16SGAYY324 pKa = 9.89VPVRR328 pKa = 11.84TCGHH332 pKa = 5.13YY333 pKa = 9.99WEE335 pKa = 5.04VTPGWSIEE343 pKa = 3.89EE344 pKa = 3.94ARR346 pKa = 11.84DD347 pKa = 3.56FFTLKK352 pKa = 8.77VTRR355 pKa = 11.84FLDD358 pKa = 3.12IAEE361 pKa = 5.18IIDD364 pKa = 4.43DD365 pKa = 4.43EE366 pKa = 5.33DD367 pKa = 4.52GQGHH371 pKa = 7.15PGAAAAADD379 pKa = 3.76SQEE382 pKa = 4.04EE383 pKa = 4.09HH384 pKa = 6.9RR385 pKa = 11.84SIYY388 pKa = 10.0HH389 pKa = 5.89HH390 pKa = 6.96
MM1 pKa = 8.05SDD3 pKa = 2.89RR4 pKa = 11.84TKK6 pKa = 11.08AKK8 pKa = 9.76RR9 pKa = 11.84WCFTINNPFNVDD21 pKa = 4.32PIIASPGAEE30 pKa = 4.7DD31 pKa = 5.73DD32 pKa = 3.69PDD34 pKa = 3.82EE35 pKa = 4.59PVVIDD40 pKa = 3.52LRR42 pKa = 11.84YY43 pKa = 9.13PFWYY47 pKa = 8.75HH48 pKa = 5.81HH49 pKa = 6.85AMLAVFKK56 pKa = 10.62VEE58 pKa = 3.94YY59 pKa = 9.55MVAQIEE65 pKa = 4.3KK66 pKa = 10.02GEE68 pKa = 4.24NGTPHH73 pKa = 5.85WQGFIIFKK81 pKa = 9.36DD82 pKa = 3.62QKK84 pKa = 10.13RR85 pKa = 11.84LQWLKK90 pKa = 11.02DD91 pKa = 3.52HH92 pKa = 7.0LDD94 pKa = 3.69KK95 pKa = 10.86RR96 pKa = 11.84AHH98 pKa = 5.89WEE100 pKa = 3.89VARR103 pKa = 11.84GTNQQASEE111 pKa = 4.33YY112 pKa = 8.85CKK114 pKa = 10.4KK115 pKa = 10.84DD116 pKa = 3.19EE117 pKa = 4.62TYY119 pKa = 10.83DD120 pKa = 3.64PSVIPADD127 pKa = 3.4MKK129 pKa = 10.35IRR131 pKa = 11.84GRR133 pKa = 11.84FEE135 pKa = 3.76WGSMPEE141 pKa = 3.88RR142 pKa = 11.84NVPKK146 pKa = 10.14KK147 pKa = 10.38DD148 pKa = 3.41EE149 pKa = 4.21RR150 pKa = 11.84LKK152 pKa = 11.1LAADD156 pKa = 3.79EE157 pKa = 5.07LDD159 pKa = 4.08TIKK162 pKa = 10.96DD163 pKa = 3.72GYY165 pKa = 10.09KK166 pKa = 9.89RR167 pKa = 11.84PAEE170 pKa = 4.18IDD172 pKa = 3.56SLTLMQCGFIPAYY185 pKa = 9.82KK186 pKa = 10.23EE187 pKa = 3.55LTADD191 pKa = 2.64ILGPYY196 pKa = 9.07RR197 pKa = 11.84PKK199 pKa = 10.82LQIITMIGPPGTGKK213 pKa = 10.4SYY215 pKa = 10.78AIQQYY220 pKa = 8.88FPNHH224 pKa = 5.6GRR226 pKa = 11.84CICGNNGVWFQNPTAPVMVFEE247 pKa = 4.71EE248 pKa = 4.71FCGQIQLQRR257 pKa = 11.84MLQYY261 pKa = 11.0LDD263 pKa = 4.6PYY265 pKa = 10.57PLALEE270 pKa = 4.65VKK272 pKa = 9.95GGMRR276 pKa = 11.84PAMYY280 pKa = 7.83EE281 pKa = 3.97TVIITSNTRR290 pKa = 11.84PDD292 pKa = 2.63GWYY295 pKa = 9.09TGDD298 pKa = 3.87QAGQPGKK305 pKa = 9.5RR306 pKa = 11.84TDD308 pKa = 3.44ALLALWDD315 pKa = 3.81RR316 pKa = 11.84LGFEE320 pKa = 4.16SGAYY324 pKa = 9.89VPVRR328 pKa = 11.84TCGHH332 pKa = 5.13YY333 pKa = 9.99WEE335 pKa = 5.04VTPGWSIEE343 pKa = 3.89EE344 pKa = 3.94ARR346 pKa = 11.84DD347 pKa = 3.56FFTLKK352 pKa = 8.77VTRR355 pKa = 11.84FLDD358 pKa = 3.12IAEE361 pKa = 5.18IIDD364 pKa = 4.43DD365 pKa = 4.43EE366 pKa = 5.33DD367 pKa = 4.52GQGHH371 pKa = 7.15PGAAAAADD379 pKa = 3.76SQEE382 pKa = 4.04EE383 pKa = 4.09HH384 pKa = 6.9RR385 pKa = 11.84SIYY388 pKa = 10.0HH389 pKa = 5.89HH390 pKa = 6.96
Molecular weight: 44.58 kDa
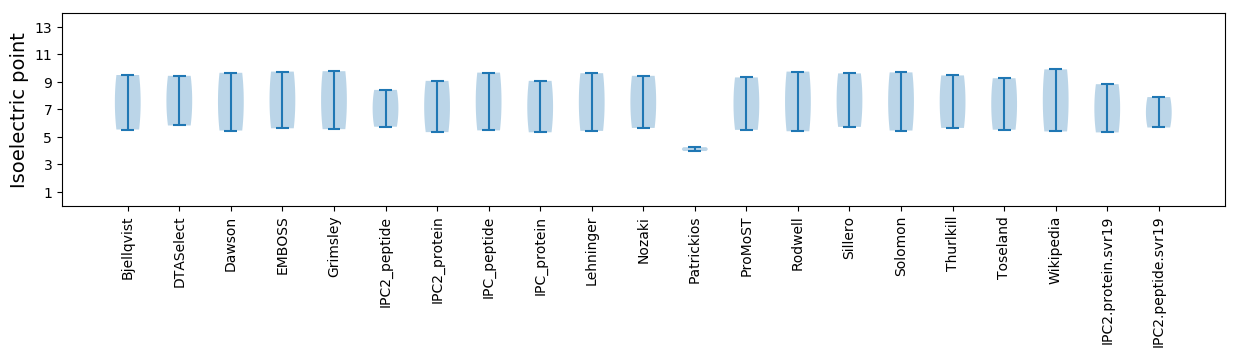
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A160HWJ0|A0A160HWJ0_9VIRU Putative capsid protein OS=Bovine faeces associated circular DNA virus 1 OX=1843764 PE=4 SV=1
MM1 pKa = 7.66PSGYY5 pKa = 9.71RR6 pKa = 11.84YY7 pKa = 9.42RR8 pKa = 11.84RR9 pKa = 11.84SYY11 pKa = 8.18YY12 pKa = 9.79GRR14 pKa = 11.84YY15 pKa = 8.33RR16 pKa = 11.84RR17 pKa = 11.84YY18 pKa = 8.91RR19 pKa = 11.84RR20 pKa = 11.84AYY22 pKa = 7.76RR23 pKa = 11.84SLYY26 pKa = 9.0RR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 9.21GRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84YY34 pKa = 9.98SYY36 pKa = 11.13ARR38 pKa = 11.84RR39 pKa = 11.84FANGSSASQVRR50 pKa = 11.84IKK52 pKa = 11.14VPVSVAYY59 pKa = 10.38AMYY62 pKa = 9.48PVTAAGSQAVCIAHH76 pKa = 7.48PFRR79 pKa = 11.84AFTSSRR85 pKa = 11.84KK86 pKa = 8.87WSPLGTKK93 pKa = 10.22LYY95 pKa = 10.03QAYY98 pKa = 10.23ANLYY102 pKa = 10.21EE103 pKa = 4.1EE104 pKa = 4.98VKK106 pKa = 10.99CIGAKK111 pKa = 9.72VQVSVCTPLGTSSIPALEE129 pKa = 5.21VYY131 pKa = 7.57TAWDD135 pKa = 3.46RR136 pKa = 11.84RR137 pKa = 11.84TGTSEE142 pKa = 3.69NGSLNSEE149 pKa = 4.37DD150 pKa = 3.36LAGYY154 pKa = 8.61GSCQQFTAVSNSVAKK169 pKa = 9.85FVRR172 pKa = 11.84SCYY175 pKa = 10.69ARR177 pKa = 11.84DD178 pKa = 3.77LMEE181 pKa = 4.28KK182 pKa = 9.94AQWHH186 pKa = 6.73DD187 pKa = 3.24SSLAEE192 pKa = 4.16STSVSDD198 pKa = 4.07EE199 pKa = 3.92AFEE202 pKa = 4.72AAGANPNFFCPGLYY216 pKa = 10.37VGFKK220 pKa = 10.14TPGLADD226 pKa = 3.63DD227 pKa = 3.98QVRR230 pKa = 11.84EE231 pKa = 3.95VDD233 pKa = 3.08IMINAKK239 pKa = 9.86YY240 pKa = 10.44YY241 pKa = 10.13YY242 pKa = 10.26VFRR245 pKa = 11.84NPGFGASAASKK256 pKa = 8.97SAVRR260 pKa = 11.84AVEE263 pKa = 3.76PVMRR267 pKa = 11.84ASAPEE272 pKa = 3.97FEE274 pKa = 5.75VEE276 pKa = 4.84DD277 pKa = 4.37LDD279 pKa = 4.84LAGDD283 pKa = 3.95EE284 pKa = 5.19DD285 pKa = 4.19VQADD289 pKa = 4.47IIAQEE294 pKa = 4.11LAAKK298 pKa = 9.99RR299 pKa = 11.84ATVKK303 pKa = 9.45KK304 pKa = 7.89TATKK308 pKa = 10.18IVTPKK313 pKa = 10.4KK314 pKa = 10.44
MM1 pKa = 7.66PSGYY5 pKa = 9.71RR6 pKa = 11.84YY7 pKa = 9.42RR8 pKa = 11.84RR9 pKa = 11.84SYY11 pKa = 8.18YY12 pKa = 9.79GRR14 pKa = 11.84YY15 pKa = 8.33RR16 pKa = 11.84RR17 pKa = 11.84YY18 pKa = 8.91RR19 pKa = 11.84RR20 pKa = 11.84AYY22 pKa = 7.76RR23 pKa = 11.84SLYY26 pKa = 9.0RR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 9.21GRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84YY34 pKa = 9.98SYY36 pKa = 11.13ARR38 pKa = 11.84RR39 pKa = 11.84FANGSSASQVRR50 pKa = 11.84IKK52 pKa = 11.14VPVSVAYY59 pKa = 10.38AMYY62 pKa = 9.48PVTAAGSQAVCIAHH76 pKa = 7.48PFRR79 pKa = 11.84AFTSSRR85 pKa = 11.84KK86 pKa = 8.87WSPLGTKK93 pKa = 10.22LYY95 pKa = 10.03QAYY98 pKa = 10.23ANLYY102 pKa = 10.21EE103 pKa = 4.1EE104 pKa = 4.98VKK106 pKa = 10.99CIGAKK111 pKa = 9.72VQVSVCTPLGTSSIPALEE129 pKa = 5.21VYY131 pKa = 7.57TAWDD135 pKa = 3.46RR136 pKa = 11.84RR137 pKa = 11.84TGTSEE142 pKa = 3.69NGSLNSEE149 pKa = 4.37DD150 pKa = 3.36LAGYY154 pKa = 8.61GSCQQFTAVSNSVAKK169 pKa = 9.85FVRR172 pKa = 11.84SCYY175 pKa = 10.69ARR177 pKa = 11.84DD178 pKa = 3.77LMEE181 pKa = 4.28KK182 pKa = 9.94AQWHH186 pKa = 6.73DD187 pKa = 3.24SSLAEE192 pKa = 4.16STSVSDD198 pKa = 4.07EE199 pKa = 3.92AFEE202 pKa = 4.72AAGANPNFFCPGLYY216 pKa = 10.37VGFKK220 pKa = 10.14TPGLADD226 pKa = 3.63DD227 pKa = 3.98QVRR230 pKa = 11.84EE231 pKa = 3.95VDD233 pKa = 3.08IMINAKK239 pKa = 9.86YY240 pKa = 10.44YY241 pKa = 10.13YY242 pKa = 10.26VFRR245 pKa = 11.84NPGFGASAASKK256 pKa = 8.97SAVRR260 pKa = 11.84AVEE263 pKa = 3.76PVMRR267 pKa = 11.84ASAPEE272 pKa = 3.97FEE274 pKa = 5.75VEE276 pKa = 4.84DD277 pKa = 4.37LDD279 pKa = 4.84LAGDD283 pKa = 3.95EE284 pKa = 5.19DD285 pKa = 4.19VQADD289 pKa = 4.47IIAQEE294 pKa = 4.11LAAKK298 pKa = 9.99RR299 pKa = 11.84ATVKK303 pKa = 9.45KK304 pKa = 7.89TATKK308 pKa = 10.18IVTPKK313 pKa = 10.4KK314 pKa = 10.44
Molecular weight: 34.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
704 |
314 |
390 |
352.0 |
39.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.227 ± 1.891 | 1.847 ± 0.039 |
5.966 ± 1.097 | 5.966 ± 0.523 |
3.977 ± 0.093 | 6.96 ± 0.546 |
1.847 ± 0.727 | 4.972 ± 1.265 |
5.398 ± 0.182 | 5.54 ± 0.458 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.273 ± 0.409 | 2.841 ± 0.015 |
5.966 ± 0.905 | 4.119 ± 0.561 |
7.102 ± 1.09 | 6.108 ± 2.261 |
4.972 ± 0.117 | 6.25 ± 1.219 |
1.989 ± 0.621 | 5.682 ± 0.987 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
