
Changjiang tombus-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.71
Get precalculated fractions of proteins
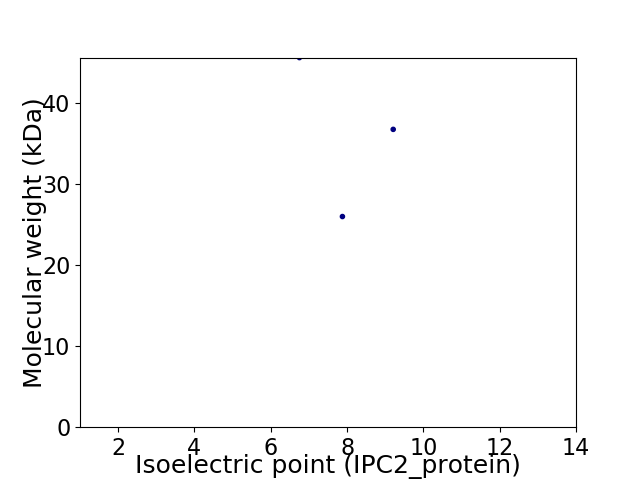
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG85|A0A1L3KG85_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 5 OX=1922819 PE=4 SV=1
MM1 pKa = 6.4YY2 pKa = 10.08QGRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.85EE8 pKa = 4.03IYY10 pKa = 9.67QRR12 pKa = 11.84AADD15 pKa = 3.96SLASRR20 pKa = 11.84PLVKK24 pKa = 10.18SDD26 pKa = 3.45AFMSTFLKK34 pKa = 10.71CEE36 pKa = 4.4KK37 pKa = 9.78IDD39 pKa = 4.01FSSKK43 pKa = 9.73PDD45 pKa = 3.62PAPRR49 pKa = 11.84VIQPRR54 pKa = 11.84TPRR57 pKa = 11.84YY58 pKa = 7.96NVEE61 pKa = 3.55VGRR64 pKa = 11.84YY65 pKa = 7.84LKK67 pKa = 10.35PLEE70 pKa = 3.89KK71 pKa = 10.0RR72 pKa = 11.84ICRR75 pKa = 11.84GIAEE79 pKa = 4.12IWGGDD84 pKa = 3.66TILKK88 pKa = 9.73GKK90 pKa = 9.68NAEE93 pKa = 3.95QSAVSLRR100 pKa = 11.84QMWDD104 pKa = 2.98QFDD107 pKa = 3.75EE108 pKa = 4.58PVAVGIDD115 pKa = 3.21ATRR118 pKa = 11.84FDD120 pKa = 3.33QHH122 pKa = 7.02VSYY125 pKa = 10.62EE126 pKa = 4.05ALEE129 pKa = 4.47WEE131 pKa = 4.3HH132 pKa = 7.06SVYY135 pKa = 10.1MLCFPPHH142 pKa = 5.89TRR144 pKa = 11.84PKK146 pKa = 10.63LEE148 pKa = 4.77RR149 pKa = 11.84LLKK152 pKa = 8.72MQLVNRR158 pKa = 11.84GYY160 pKa = 11.34ARR162 pKa = 11.84LEE164 pKa = 4.05DD165 pKa = 4.14LEE167 pKa = 4.94LMYY170 pKa = 10.41EE171 pKa = 4.06VRR173 pKa = 11.84GRR175 pKa = 11.84RR176 pKa = 11.84MSGDD180 pKa = 3.3MNTGMGNCLLMCAMIHH196 pKa = 6.94WITAEE201 pKa = 3.7MGVRR205 pKa = 11.84CRR207 pKa = 11.84LANNGDD213 pKa = 3.75DD214 pKa = 4.14CVLILEE220 pKa = 4.64KK221 pKa = 10.34RR222 pKa = 11.84DD223 pKa = 3.47LGKK226 pKa = 10.58LGRR229 pKa = 11.84LGPLALDD236 pKa = 3.85FGFVLEE242 pKa = 5.02IEE244 pKa = 4.53PEE246 pKa = 3.85VDD248 pKa = 3.52VFEE251 pKa = 5.33QICFCQNHH259 pKa = 5.93PVWGGHH265 pKa = 4.52SWVMCRR271 pKa = 11.84DD272 pKa = 3.24PRR274 pKa = 11.84KK275 pKa = 10.24CVDD278 pKa = 3.34KK279 pKa = 11.36DD280 pKa = 3.57LVTVLDD286 pKa = 4.21LGNIKK291 pKa = 9.81SARR294 pKa = 11.84KK295 pKa = 6.53WCHH298 pKa = 6.13AIGTGGLAMAGGLPVLGSFYY318 pKa = 11.49GMLLRR323 pKa = 11.84HH324 pKa = 5.73ATTGKK329 pKa = 10.23VDD331 pKa = 3.36AHH333 pKa = 6.38PWLEE337 pKa = 3.89NGFAMMAKK345 pKa = 10.02GMARR349 pKa = 11.84HH350 pKa = 6.09SDD352 pKa = 3.34VVTPEE357 pKa = 3.67SRR359 pKa = 11.84ASFFKK364 pKa = 11.03AFDD367 pKa = 3.44ITPDD371 pKa = 3.32QQEE374 pKa = 4.67SIEE377 pKa = 4.17SSYY380 pKa = 11.27ATMLLNLSAGVSEE393 pKa = 5.57SPLACYY399 pKa = 10.08GKK401 pKa = 9.55IWQQ404 pKa = 4.11
MM1 pKa = 6.4YY2 pKa = 10.08QGRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.85EE8 pKa = 4.03IYY10 pKa = 9.67QRR12 pKa = 11.84AADD15 pKa = 3.96SLASRR20 pKa = 11.84PLVKK24 pKa = 10.18SDD26 pKa = 3.45AFMSTFLKK34 pKa = 10.71CEE36 pKa = 4.4KK37 pKa = 9.78IDD39 pKa = 4.01FSSKK43 pKa = 9.73PDD45 pKa = 3.62PAPRR49 pKa = 11.84VIQPRR54 pKa = 11.84TPRR57 pKa = 11.84YY58 pKa = 7.96NVEE61 pKa = 3.55VGRR64 pKa = 11.84YY65 pKa = 7.84LKK67 pKa = 10.35PLEE70 pKa = 3.89KK71 pKa = 10.0RR72 pKa = 11.84ICRR75 pKa = 11.84GIAEE79 pKa = 4.12IWGGDD84 pKa = 3.66TILKK88 pKa = 9.73GKK90 pKa = 9.68NAEE93 pKa = 3.95QSAVSLRR100 pKa = 11.84QMWDD104 pKa = 2.98QFDD107 pKa = 3.75EE108 pKa = 4.58PVAVGIDD115 pKa = 3.21ATRR118 pKa = 11.84FDD120 pKa = 3.33QHH122 pKa = 7.02VSYY125 pKa = 10.62EE126 pKa = 4.05ALEE129 pKa = 4.47WEE131 pKa = 4.3HH132 pKa = 7.06SVYY135 pKa = 10.1MLCFPPHH142 pKa = 5.89TRR144 pKa = 11.84PKK146 pKa = 10.63LEE148 pKa = 4.77RR149 pKa = 11.84LLKK152 pKa = 8.72MQLVNRR158 pKa = 11.84GYY160 pKa = 11.34ARR162 pKa = 11.84LEE164 pKa = 4.05DD165 pKa = 4.14LEE167 pKa = 4.94LMYY170 pKa = 10.41EE171 pKa = 4.06VRR173 pKa = 11.84GRR175 pKa = 11.84RR176 pKa = 11.84MSGDD180 pKa = 3.3MNTGMGNCLLMCAMIHH196 pKa = 6.94WITAEE201 pKa = 3.7MGVRR205 pKa = 11.84CRR207 pKa = 11.84LANNGDD213 pKa = 3.75DD214 pKa = 4.14CVLILEE220 pKa = 4.64KK221 pKa = 10.34RR222 pKa = 11.84DD223 pKa = 3.47LGKK226 pKa = 10.58LGRR229 pKa = 11.84LGPLALDD236 pKa = 3.85FGFVLEE242 pKa = 5.02IEE244 pKa = 4.53PEE246 pKa = 3.85VDD248 pKa = 3.52VFEE251 pKa = 5.33QICFCQNHH259 pKa = 5.93PVWGGHH265 pKa = 4.52SWVMCRR271 pKa = 11.84DD272 pKa = 3.24PRR274 pKa = 11.84KK275 pKa = 10.24CVDD278 pKa = 3.34KK279 pKa = 11.36DD280 pKa = 3.57LVTVLDD286 pKa = 4.21LGNIKK291 pKa = 9.81SARR294 pKa = 11.84KK295 pKa = 6.53WCHH298 pKa = 6.13AIGTGGLAMAGGLPVLGSFYY318 pKa = 11.49GMLLRR323 pKa = 11.84HH324 pKa = 5.73ATTGKK329 pKa = 10.23VDD331 pKa = 3.36AHH333 pKa = 6.38PWLEE337 pKa = 3.89NGFAMMAKK345 pKa = 10.02GMARR349 pKa = 11.84HH350 pKa = 6.09SDD352 pKa = 3.34VVTPEE357 pKa = 3.67SRR359 pKa = 11.84ASFFKK364 pKa = 11.03AFDD367 pKa = 3.44ITPDD371 pKa = 3.32QQEE374 pKa = 4.67SIEE377 pKa = 4.17SSYY380 pKa = 11.27ATMLLNLSAGVSEE393 pKa = 5.57SPLACYY399 pKa = 10.08GKK401 pKa = 9.55IWQQ404 pKa = 4.11
Molecular weight: 45.55 kDa
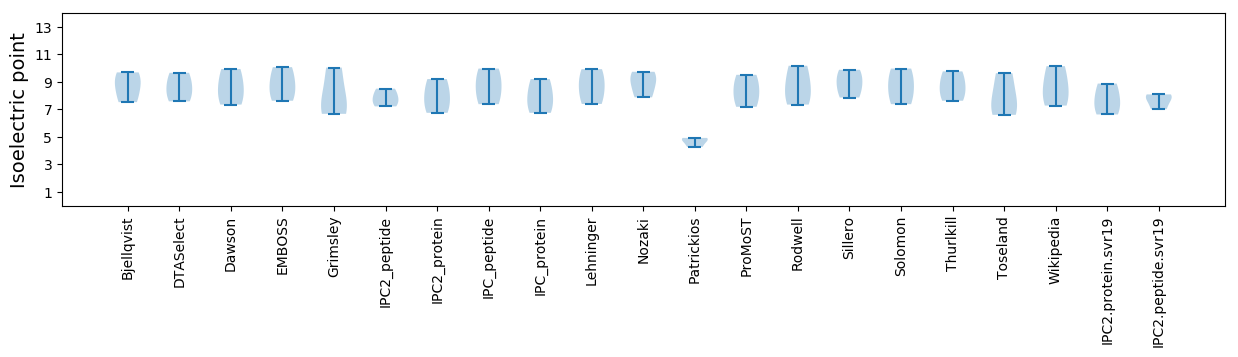
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFT1|A0A1L3KFT1_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 5 OX=1922819 PE=4 SV=1
MM1 pKa = 7.82AKK3 pKa = 9.19TPKK6 pKa = 10.45NKK8 pKa = 10.03GKK10 pKa = 9.04SQSVRR15 pKa = 11.84VLVTGGRR22 pKa = 11.84KK23 pKa = 9.26KK24 pKa = 10.29IKK26 pKa = 10.24RR27 pKa = 11.84KK28 pKa = 8.29ATAGNRR34 pKa = 11.84MGMSTLPARR43 pKa = 11.84WMRR46 pKa = 11.84LLTDD50 pKa = 3.86PCLADD55 pKa = 3.58LTYY58 pKa = 10.29PCYY61 pKa = 10.63GGTDD65 pKa = 2.67AGYY68 pKa = 10.59LVRR71 pKa = 11.84TVDD74 pKa = 3.53TFSPTIGGSFTVGSQVIADD93 pKa = 5.36GIYY96 pKa = 8.96TWSPWNLSATTGTRR110 pKa = 11.84SSFWTSGDD118 pKa = 3.37PLTPSSAGFNGNFISTAAVRR138 pKa = 11.84SYY140 pKa = 11.18RR141 pKa = 11.84PVASCLKK148 pKa = 8.63WVPIGPIGGRR158 pKa = 11.84SGLIASGYY166 pKa = 9.05SPGVVATPSVGVGNYY181 pKa = 6.81VTLAQRR187 pKa = 11.84KK188 pKa = 6.04ATNGTEE194 pKa = 3.65MHH196 pKa = 7.14EE197 pKa = 4.54INWLPTSHH205 pKa = 7.46DD206 pKa = 3.63EE207 pKa = 4.28EE208 pKa = 4.72FTTVEE213 pKa = 4.19QANSTSVGTVFMALKK228 pKa = 10.58GVDD231 pKa = 3.7AVAKK235 pKa = 9.93SSTSATLNGYY245 pKa = 8.86FEE247 pKa = 4.57ITTVWEE253 pKa = 3.99WTPATTTGLCVDD265 pKa = 4.46PRR267 pKa = 11.84TPSPYY272 pKa = 9.36TSQTVLSMFGDD283 pKa = 3.66IKK285 pKa = 11.07NALFAGAHH293 pKa = 4.69STAVGAAGQFGRR305 pKa = 11.84KK306 pKa = 8.01AVGMIGGVASSAAMAAIDD324 pKa = 3.7YY325 pKa = 10.1AGRR328 pKa = 11.84SFMGVANNPPRR339 pKa = 11.84YY340 pKa = 9.21RR341 pKa = 11.84GNSVPLIGYY350 pKa = 8.49
MM1 pKa = 7.82AKK3 pKa = 9.19TPKK6 pKa = 10.45NKK8 pKa = 10.03GKK10 pKa = 9.04SQSVRR15 pKa = 11.84VLVTGGRR22 pKa = 11.84KK23 pKa = 9.26KK24 pKa = 10.29IKK26 pKa = 10.24RR27 pKa = 11.84KK28 pKa = 8.29ATAGNRR34 pKa = 11.84MGMSTLPARR43 pKa = 11.84WMRR46 pKa = 11.84LLTDD50 pKa = 3.86PCLADD55 pKa = 3.58LTYY58 pKa = 10.29PCYY61 pKa = 10.63GGTDD65 pKa = 2.67AGYY68 pKa = 10.59LVRR71 pKa = 11.84TVDD74 pKa = 3.53TFSPTIGGSFTVGSQVIADD93 pKa = 5.36GIYY96 pKa = 8.96TWSPWNLSATTGTRR110 pKa = 11.84SSFWTSGDD118 pKa = 3.37PLTPSSAGFNGNFISTAAVRR138 pKa = 11.84SYY140 pKa = 11.18RR141 pKa = 11.84PVASCLKK148 pKa = 8.63WVPIGPIGGRR158 pKa = 11.84SGLIASGYY166 pKa = 9.05SPGVVATPSVGVGNYY181 pKa = 6.81VTLAQRR187 pKa = 11.84KK188 pKa = 6.04ATNGTEE194 pKa = 3.65MHH196 pKa = 7.14EE197 pKa = 4.54INWLPTSHH205 pKa = 7.46DD206 pKa = 3.63EE207 pKa = 4.28EE208 pKa = 4.72FTTVEE213 pKa = 4.19QANSTSVGTVFMALKK228 pKa = 10.58GVDD231 pKa = 3.7AVAKK235 pKa = 9.93SSTSATLNGYY245 pKa = 8.86FEE247 pKa = 4.57ITTVWEE253 pKa = 3.99WTPATTTGLCVDD265 pKa = 4.46PRR267 pKa = 11.84TPSPYY272 pKa = 9.36TSQTVLSMFGDD283 pKa = 3.66IKK285 pKa = 11.07NALFAGAHH293 pKa = 4.69STAVGAAGQFGRR305 pKa = 11.84KK306 pKa = 8.01AVGMIGGVASSAAMAAIDD324 pKa = 3.7YY325 pKa = 10.1AGRR328 pKa = 11.84SFMGVANNPPRR339 pKa = 11.84YY340 pKa = 9.21RR341 pKa = 11.84GNSVPLIGYY350 pKa = 8.49
Molecular weight: 36.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
990 |
236 |
404 |
330.0 |
36.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.495 ± 1.092 | 2.222 ± 0.534 |
4.949 ± 0.743 | 4.747 ± 1.126 |
3.434 ± 0.169 | 9.091 ± 1.086 |
1.616 ± 0.438 | 3.838 ± 0.43 |
4.747 ± 0.311 | 8.283 ± 1.178 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.242 ± 0.576 | 3.03 ± 0.405 |
4.949 ± 0.523 | 2.121 ± 0.581 |
6.768 ± 0.833 | 6.97 ± 1.12 |
6.768 ± 1.947 | 7.98 ± 0.941 |
1.919 ± 0.33 | 2.828 ± 0.267 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
