
Oryzias javanicus (Javanese ricefish) (Aplocheilus javanicus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri;
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
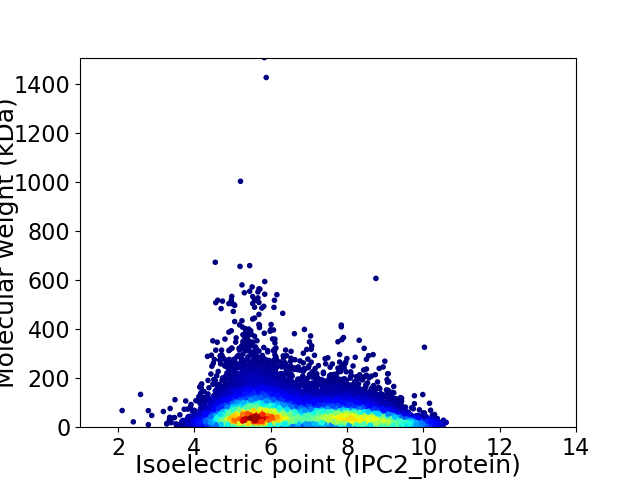
Virtual 2D-PAGE plot for 21400 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A437CXI4|A0A437CXI4_ORYJA Uncharacterized protein OS=Oryzias javanicus OX=123683 GN=OJAV_G00101990 PE=3 SV=1
MM1 pKa = 7.08QPSALRR7 pKa = 11.84ALCSALLLLTVVPSVCSQSDD27 pKa = 3.66CSQADD32 pKa = 3.52SCDD35 pKa = 3.65LCVGDD40 pKa = 5.83SMLNLTGCVWRR51 pKa = 11.84LCPNGNDD58 pKa = 3.04TGMCVTDD65 pKa = 4.75GGDD68 pKa = 4.16SNDD71 pKa = 3.38TGMNCSWTRR80 pKa = 11.84VSEE83 pKa = 4.16LCTVVEE89 pKa = 4.51NVATGGEE96 pKa = 4.68GDD98 pKa = 4.03TGDD101 pKa = 4.78ASDD104 pKa = 3.9TTSSPEE110 pKa = 3.53FSQAKK115 pKa = 9.54FDD117 pKa = 3.55MSSFIGGIILVLSLQAGGFFAMRR140 pKa = 11.84FLKK143 pKa = 10.72SKK145 pKa = 9.46EE146 pKa = 3.68QSNYY150 pKa = 10.84DD151 pKa = 4.46PIEE154 pKa = 3.99QPQQ157 pKa = 3.0
MM1 pKa = 7.08QPSALRR7 pKa = 11.84ALCSALLLLTVVPSVCSQSDD27 pKa = 3.66CSQADD32 pKa = 3.52SCDD35 pKa = 3.65LCVGDD40 pKa = 5.83SMLNLTGCVWRR51 pKa = 11.84LCPNGNDD58 pKa = 3.04TGMCVTDD65 pKa = 4.75GGDD68 pKa = 4.16SNDD71 pKa = 3.38TGMNCSWTRR80 pKa = 11.84VSEE83 pKa = 4.16LCTVVEE89 pKa = 4.51NVATGGEE96 pKa = 4.68GDD98 pKa = 4.03TGDD101 pKa = 4.78ASDD104 pKa = 3.9TTSSPEE110 pKa = 3.53FSQAKK115 pKa = 9.54FDD117 pKa = 3.55MSSFIGGIILVLSLQAGGFFAMRR140 pKa = 11.84FLKK143 pKa = 10.72SKK145 pKa = 9.46EE146 pKa = 3.68QSNYY150 pKa = 10.84DD151 pKa = 4.46PIEE154 pKa = 3.99QPQQ157 pKa = 3.0
Molecular weight: 16.53 kDa
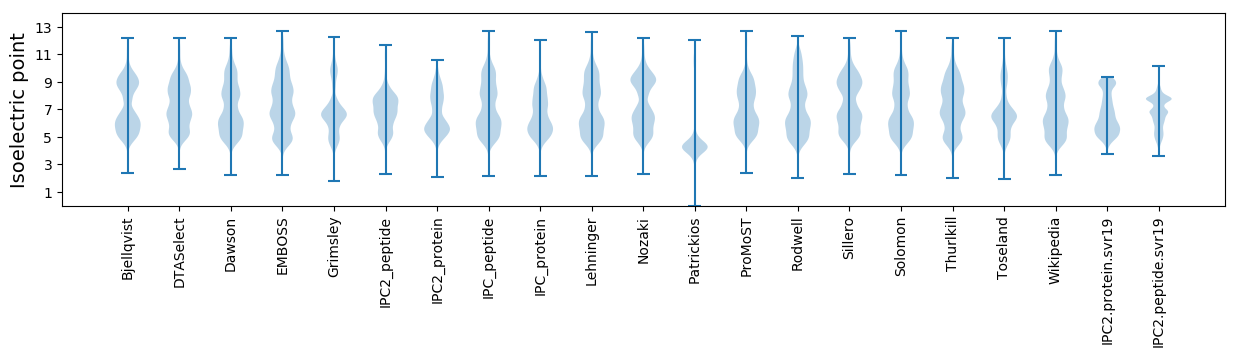
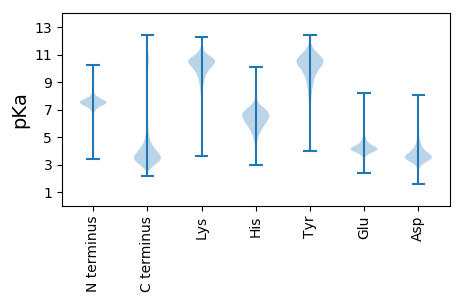
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A437CM98|A0A437CM98_ORYJA Uncharacterized protein OS=Oryzias javanicus OX=123683 GN=OJAV_G00163900 PE=4 SV=1
MM1 pKa = 7.32EE2 pKa = 4.9FSYY5 pKa = 10.99GRR7 pKa = 11.84SFLTPTTNQRR17 pKa = 11.84LLKK20 pKa = 8.34EE21 pKa = 3.92THH23 pKa = 6.09LRR25 pKa = 11.84RR26 pKa = 11.84SGPLKK31 pKa = 9.74EE32 pKa = 4.31SPRR35 pKa = 11.84ARR37 pKa = 11.84WLISRR42 pKa = 11.84GTLNLSRR49 pKa = 11.84PPPPHH54 pKa = 6.49EE55 pKa = 4.23LPRR58 pKa = 11.84AQEE61 pKa = 4.06RR62 pKa = 11.84RR63 pKa = 11.84HH64 pKa = 5.43GAALYY69 pKa = 10.31ARR71 pKa = 11.84SAPAPEE77 pKa = 5.82LEE79 pKa = 4.27LQSNASLNRR88 pKa = 11.84ARR90 pKa = 11.84AGGRR94 pKa = 11.84GAGGSAHH101 pKa = 6.16EE102 pKa = 4.21RR103 pKa = 11.84AEE105 pKa = 4.08RR106 pKa = 11.84SQVGCRR112 pKa = 11.84EE113 pKa = 3.99LRR115 pKa = 11.84STKK118 pKa = 10.35YY119 pKa = 10.43ISDD122 pKa = 3.73GRR124 pKa = 11.84CTSINPIKK132 pKa = 10.47EE133 pKa = 4.01LVCAGEE139 pKa = 4.34CLPAQMFEE147 pKa = 3.61NWIGGRR153 pKa = 11.84RR154 pKa = 11.84YY155 pKa = 7.81WARR158 pKa = 11.84RR159 pKa = 11.84SGNDD163 pKa = 2.96DD164 pKa = 3.17WRR166 pKa = 11.84CVNDD170 pKa = 3.23KK171 pKa = 10.15TRR173 pKa = 11.84TQRR176 pKa = 11.84IQLQCQDD183 pKa = 3.43GSSRR187 pKa = 11.84TYY189 pKa = 10.45KK190 pKa = 8.86ITVVTSCKK198 pKa = 9.38CKK200 pKa = 10.21RR201 pKa = 11.84YY202 pKa = 9.78SRR204 pKa = 11.84QHH206 pKa = 5.8NEE208 pKa = 3.62SGGKK212 pKa = 9.71FEE214 pKa = 5.14EE215 pKa = 4.82PAAVPPKK222 pKa = 10.17LLHH225 pKa = 5.33KK226 pKa = 9.98HH227 pKa = 5.33KK228 pKa = 10.48SKK230 pKa = 10.56SRR232 pKa = 11.84RR233 pKa = 11.84RR234 pKa = 11.84LGKK237 pKa = 10.04SRR239 pKa = 11.84TRR241 pKa = 11.84EE242 pKa = 3.7NWHH245 pKa = 6.12DD246 pKa = 3.71TEE248 pKa = 4.38PP249 pKa = 3.93
MM1 pKa = 7.32EE2 pKa = 4.9FSYY5 pKa = 10.99GRR7 pKa = 11.84SFLTPTTNQRR17 pKa = 11.84LLKK20 pKa = 8.34EE21 pKa = 3.92THH23 pKa = 6.09LRR25 pKa = 11.84RR26 pKa = 11.84SGPLKK31 pKa = 9.74EE32 pKa = 4.31SPRR35 pKa = 11.84ARR37 pKa = 11.84WLISRR42 pKa = 11.84GTLNLSRR49 pKa = 11.84PPPPHH54 pKa = 6.49EE55 pKa = 4.23LPRR58 pKa = 11.84AQEE61 pKa = 4.06RR62 pKa = 11.84RR63 pKa = 11.84HH64 pKa = 5.43GAALYY69 pKa = 10.31ARR71 pKa = 11.84SAPAPEE77 pKa = 5.82LEE79 pKa = 4.27LQSNASLNRR88 pKa = 11.84ARR90 pKa = 11.84AGGRR94 pKa = 11.84GAGGSAHH101 pKa = 6.16EE102 pKa = 4.21RR103 pKa = 11.84AEE105 pKa = 4.08RR106 pKa = 11.84SQVGCRR112 pKa = 11.84EE113 pKa = 3.99LRR115 pKa = 11.84STKK118 pKa = 10.35YY119 pKa = 10.43ISDD122 pKa = 3.73GRR124 pKa = 11.84CTSINPIKK132 pKa = 10.47EE133 pKa = 4.01LVCAGEE139 pKa = 4.34CLPAQMFEE147 pKa = 3.61NWIGGRR153 pKa = 11.84RR154 pKa = 11.84YY155 pKa = 7.81WARR158 pKa = 11.84RR159 pKa = 11.84SGNDD163 pKa = 2.96DD164 pKa = 3.17WRR166 pKa = 11.84CVNDD170 pKa = 3.23KK171 pKa = 10.15TRR173 pKa = 11.84TQRR176 pKa = 11.84IQLQCQDD183 pKa = 3.43GSSRR187 pKa = 11.84TYY189 pKa = 10.45KK190 pKa = 8.86ITVVTSCKK198 pKa = 9.38CKK200 pKa = 10.21RR201 pKa = 11.84YY202 pKa = 9.78SRR204 pKa = 11.84QHH206 pKa = 5.8NEE208 pKa = 3.62SGGKK212 pKa = 9.71FEE214 pKa = 5.14EE215 pKa = 4.82PAAVPPKK222 pKa = 10.17LLHH225 pKa = 5.33KK226 pKa = 9.98HH227 pKa = 5.33KK228 pKa = 10.48SKK230 pKa = 10.56SRR232 pKa = 11.84RR233 pKa = 11.84RR234 pKa = 11.84LGKK237 pKa = 10.04SRR239 pKa = 11.84TRR241 pKa = 11.84EE242 pKa = 3.7NWHH245 pKa = 6.12DD246 pKa = 3.71TEE248 pKa = 4.38PP249 pKa = 3.93
Molecular weight: 28.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11474022 |
51 |
13616 |
536.2 |
59.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.747 ± 0.015 | 2.254 ± 0.014 |
5.151 ± 0.012 | 6.963 ± 0.021 |
3.633 ± 0.01 | 6.376 ± 0.022 |
2.584 ± 0.009 | 4.15 ± 0.012 |
5.612 ± 0.021 | 9.563 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.315 ± 0.008 | 3.73 ± 0.01 |
5.859 ± 0.022 | 4.761 ± 0.018 |
5.842 ± 0.014 | 8.979 ± 0.022 |
5.487 ± 0.015 | 6.267 ± 0.014 |
1.172 ± 0.005 | 2.553 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
