
Acidipila rosea
Taxonomy: cellular organisms; Bacteria; Acidobacteria; Acidobacteriia; Acidobacteriales; Acidobacteriaceae; Acidipila
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
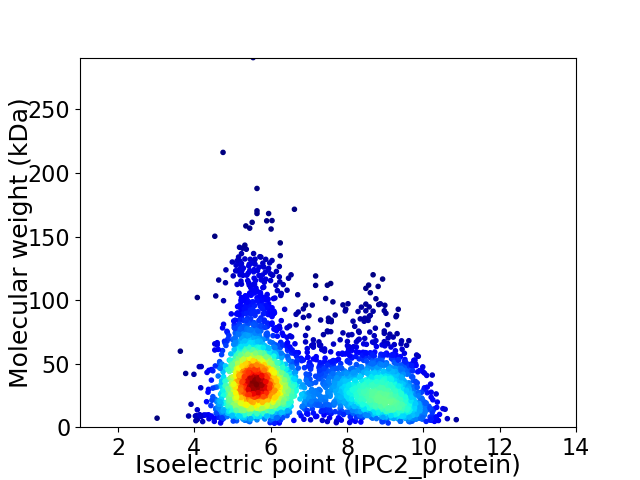
Virtual 2D-PAGE plot for 3487 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
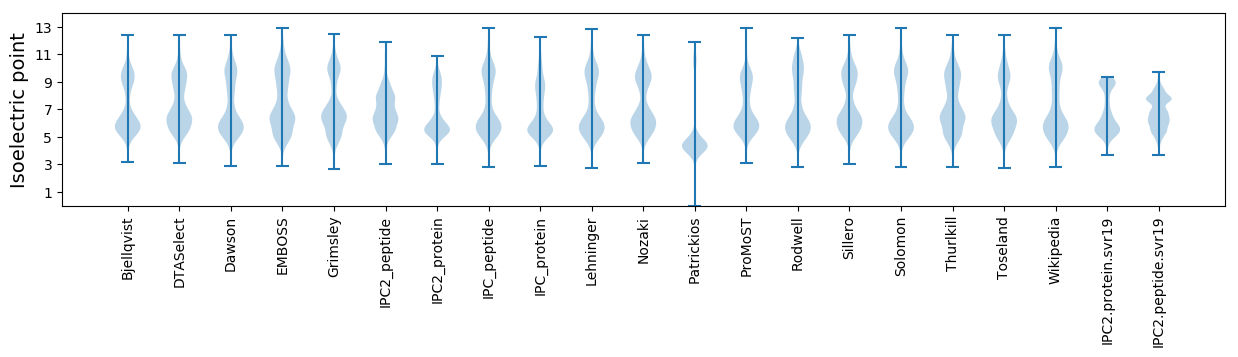
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R1LAX1|A0A4R1LAX1_9BACT Uncharacterized protein OS=Acidipila rosea OX=768535 GN=C7378_0468 PE=4 SV=1
MM1 pKa = 7.76EE2 pKa = 5.22SFSSLSQSGSANFFQAPSDD21 pKa = 3.9FADD24 pKa = 3.6TRR26 pKa = 11.84SMGTVGLSFGSPTSGTGFNVTSTVNQIVTNMQTVEE61 pKa = 4.6TPWKK65 pKa = 7.88TQLTSLQAEE74 pKa = 4.46DD75 pKa = 3.71TALTSIGTDD84 pKa = 3.52LNALSTSMQALTDD97 pKa = 3.94FQGVLAGKK105 pKa = 9.81QGSSSDD111 pKa = 3.71TNTLQLTSAATTAIAGSHH129 pKa = 6.1TIVVSQLAQTSSYY142 pKa = 11.07VSGAIANASDD152 pKa = 3.66TLAGSLTLSIGSGSSQTITLNSSDD176 pKa = 3.53NTLAGLANTINSGSYY191 pKa = 10.11GVTANVITDD200 pKa = 3.5SSGSRR205 pKa = 11.84LSLVSSTSGAGGDD218 pKa = 3.45ITLSSSMTDD227 pKa = 3.16TSNSNAAVTFTQAQAGQDD245 pKa = 3.11ALMTVDD251 pKa = 3.98GVSVTSASNTVTNAIQGVTFQLLKK275 pKa = 10.76ASPSEE280 pKa = 3.91NVQVQITNDD289 pKa = 3.36NTSVTTAVNTFVTAYY304 pKa = 10.66NKK306 pKa = 10.01VLTDD310 pKa = 4.59LNTQEE315 pKa = 5.28GNTSSGTPEE324 pKa = 3.99PLFGNPVLANLQQQLQAALTFTQPAQAVATNTTIGTSDD362 pKa = 3.43TLAGSLSIAVGTGAAQTVAVPSSTPTLAGLASAINAANMGVTANVITSGSQSTLSLQNATSGSAGVININSSSLSDD438 pKa = 3.39TTTGQAVTFGGSLSAGVTSLTQLGITVNNDD468 pKa = 2.06GTLALNGDD476 pKa = 4.27TLNSEE481 pKa = 4.7LNSNYY486 pKa = 9.72QDD488 pKa = 2.94VVNFFQPSGNFTSFGGNFSTVMGNLGNSAPSGAVYY523 pKa = 10.47LAMQEE528 pKa = 4.0NSTTEE533 pKa = 3.93SQLNTNVSNEE543 pKa = 3.77EE544 pKa = 4.3AIINTQKK551 pKa = 10.21ATLTTEE557 pKa = 4.59LNQANFTLTQIPQQLNYY574 pKa = 9.99MNEE577 pKa = 4.26LYY579 pKa = 10.8SAITGYY585 pKa = 8.17NTYY588 pKa = 10.57HH589 pKa = 6.97PP590 pKa = 4.67
MM1 pKa = 7.76EE2 pKa = 5.22SFSSLSQSGSANFFQAPSDD21 pKa = 3.9FADD24 pKa = 3.6TRR26 pKa = 11.84SMGTVGLSFGSPTSGTGFNVTSTVNQIVTNMQTVEE61 pKa = 4.6TPWKK65 pKa = 7.88TQLTSLQAEE74 pKa = 4.46DD75 pKa = 3.71TALTSIGTDD84 pKa = 3.52LNALSTSMQALTDD97 pKa = 3.94FQGVLAGKK105 pKa = 9.81QGSSSDD111 pKa = 3.71TNTLQLTSAATTAIAGSHH129 pKa = 6.1TIVVSQLAQTSSYY142 pKa = 11.07VSGAIANASDD152 pKa = 3.66TLAGSLTLSIGSGSSQTITLNSSDD176 pKa = 3.53NTLAGLANTINSGSYY191 pKa = 10.11GVTANVITDD200 pKa = 3.5SSGSRR205 pKa = 11.84LSLVSSTSGAGGDD218 pKa = 3.45ITLSSSMTDD227 pKa = 3.16TSNSNAAVTFTQAQAGQDD245 pKa = 3.11ALMTVDD251 pKa = 3.98GVSVTSASNTVTNAIQGVTFQLLKK275 pKa = 10.76ASPSEE280 pKa = 3.91NVQVQITNDD289 pKa = 3.36NTSVTTAVNTFVTAYY304 pKa = 10.66NKK306 pKa = 10.01VLTDD310 pKa = 4.59LNTQEE315 pKa = 5.28GNTSSGTPEE324 pKa = 3.99PLFGNPVLANLQQQLQAALTFTQPAQAVATNTTIGTSDD362 pKa = 3.43TLAGSLSIAVGTGAAQTVAVPSSTPTLAGLASAINAANMGVTANVITSGSQSTLSLQNATSGSAGVININSSSLSDD438 pKa = 3.39TTTGQAVTFGGSLSAGVTSLTQLGITVNNDD468 pKa = 2.06GTLALNGDD476 pKa = 4.27TLNSEE481 pKa = 4.7LNSNYY486 pKa = 9.72QDD488 pKa = 2.94VVNFFQPSGNFTSFGGNFSTVMGNLGNSAPSGAVYY523 pKa = 10.47LAMQEE528 pKa = 4.0NSTTEE533 pKa = 3.93SQLNTNVSNEE543 pKa = 3.77EE544 pKa = 4.3AIINTQKK551 pKa = 10.21ATLTTEE557 pKa = 4.59LNQANFTLTQIPQQLNYY574 pKa = 9.99MNEE577 pKa = 4.26LYY579 pKa = 10.8SAITGYY585 pKa = 8.17NTYY588 pKa = 10.57HH589 pKa = 6.97PP590 pKa = 4.67
Molecular weight: 59.79 kDa
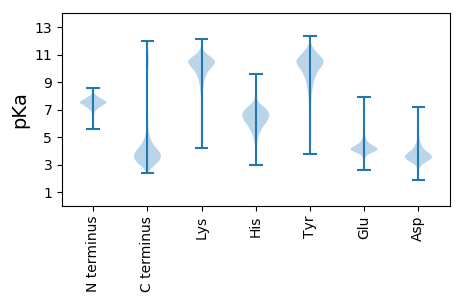
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R1L3X6|A0A4R1L3X6_9BACT Nucleotide-binding universal stress UspA family protein OS=Acidipila rosea OX=768535 GN=C7378_2345 PE=3 SV=1
MM1 pKa = 7.89PEE3 pKa = 3.96LPEE6 pKa = 4.22VEE8 pKa = 4.44TVANGVHH15 pKa = 5.41EE16 pKa = 5.39RR17 pKa = 11.84IHH19 pKa = 6.29GSRR22 pKa = 11.84VEE24 pKa = 4.04QAWFGSKK31 pKa = 10.25PEE33 pKa = 4.14PFKK36 pKa = 10.91TPPAEE41 pKa = 3.66MAAAITGATIDD52 pKa = 3.4RR53 pKa = 11.84VHH55 pKa = 6.88RR56 pKa = 11.84VGKK59 pKa = 10.01HH60 pKa = 3.95IVFDD64 pKa = 3.92LTPAPPASGSLQWIVHH80 pKa = 6.25LGMTGRR86 pKa = 11.84LLVSTPDD93 pKa = 3.58TPVPAHH99 pKa = 5.32THH101 pKa = 5.94ALLTLDD107 pKa = 3.74SGRR110 pKa = 11.84EE111 pKa = 3.87LRR113 pKa = 11.84FVDD116 pKa = 3.32ARR118 pKa = 11.84RR119 pKa = 11.84FGRR122 pKa = 11.84LALQRR127 pKa = 11.84TAFTGPGAEE136 pKa = 4.6PLTISADD143 pKa = 3.51DD144 pKa = 3.82FVALFRR150 pKa = 11.84GRR152 pKa = 11.84KK153 pKa = 8.77LAIKK157 pKa = 10.18SALLNQRR164 pKa = 11.84LLHH167 pKa = 5.82GVGNIYY173 pKa = 10.78ADD175 pKa = 3.41EE176 pKa = 4.3SLFRR180 pKa = 11.84AGIRR184 pKa = 11.84PTHH187 pKa = 5.37IAGRR191 pKa = 11.84LSRR194 pKa = 11.84PRR196 pKa = 11.84LQRR199 pKa = 11.84LHH201 pKa = 6.05TEE203 pKa = 3.91LRR205 pKa = 11.84AVLRR209 pKa = 11.84NAIALGGSSVSDD221 pKa = 3.57YY222 pKa = 11.52VDD224 pKa = 3.5ADD226 pKa = 3.53GVRR229 pKa = 11.84GFFQLEE235 pKa = 3.77HH236 pKa = 6.45RR237 pKa = 11.84VYY239 pKa = 10.67QRR241 pKa = 11.84GGQPCLVCGTAIRR254 pKa = 11.84RR255 pKa = 11.84TVLGGRR261 pKa = 11.84STHH264 pKa = 5.38WCPRR268 pKa = 11.84CQSS271 pKa = 3.2
MM1 pKa = 7.89PEE3 pKa = 3.96LPEE6 pKa = 4.22VEE8 pKa = 4.44TVANGVHH15 pKa = 5.41EE16 pKa = 5.39RR17 pKa = 11.84IHH19 pKa = 6.29GSRR22 pKa = 11.84VEE24 pKa = 4.04QAWFGSKK31 pKa = 10.25PEE33 pKa = 4.14PFKK36 pKa = 10.91TPPAEE41 pKa = 3.66MAAAITGATIDD52 pKa = 3.4RR53 pKa = 11.84VHH55 pKa = 6.88RR56 pKa = 11.84VGKK59 pKa = 10.01HH60 pKa = 3.95IVFDD64 pKa = 3.92LTPAPPASGSLQWIVHH80 pKa = 6.25LGMTGRR86 pKa = 11.84LLVSTPDD93 pKa = 3.58TPVPAHH99 pKa = 5.32THH101 pKa = 5.94ALLTLDD107 pKa = 3.74SGRR110 pKa = 11.84EE111 pKa = 3.87LRR113 pKa = 11.84FVDD116 pKa = 3.32ARR118 pKa = 11.84RR119 pKa = 11.84FGRR122 pKa = 11.84LALQRR127 pKa = 11.84TAFTGPGAEE136 pKa = 4.6PLTISADD143 pKa = 3.51DD144 pKa = 3.82FVALFRR150 pKa = 11.84GRR152 pKa = 11.84KK153 pKa = 8.77LAIKK157 pKa = 10.18SALLNQRR164 pKa = 11.84LLHH167 pKa = 5.82GVGNIYY173 pKa = 10.78ADD175 pKa = 3.41EE176 pKa = 4.3SLFRR180 pKa = 11.84AGIRR184 pKa = 11.84PTHH187 pKa = 5.37IAGRR191 pKa = 11.84LSRR194 pKa = 11.84PRR196 pKa = 11.84LQRR199 pKa = 11.84LHH201 pKa = 6.05TEE203 pKa = 3.91LRR205 pKa = 11.84AVLRR209 pKa = 11.84NAIALGGSSVSDD221 pKa = 3.57YY222 pKa = 11.52VDD224 pKa = 3.5ADD226 pKa = 3.53GVRR229 pKa = 11.84GFFQLEE235 pKa = 3.77HH236 pKa = 6.45RR237 pKa = 11.84VYY239 pKa = 10.67QRR241 pKa = 11.84GGQPCLVCGTAIRR254 pKa = 11.84RR255 pKa = 11.84TVLGGRR261 pKa = 11.84STHH264 pKa = 5.38WCPRR268 pKa = 11.84CQSS271 pKa = 3.2
Molecular weight: 29.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1276136 |
29 |
2650 |
366.0 |
40.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.004 ± 0.051 | 0.86 ± 0.013 |
4.993 ± 0.028 | 5.574 ± 0.045 |
3.838 ± 0.025 | 7.85 ± 0.045 |
2.396 ± 0.02 | 5.102 ± 0.027 |
3.614 ± 0.034 | 10.254 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.326 ± 0.017 | 3.278 ± 0.038 |
5.274 ± 0.029 | 4.077 ± 0.031 |
6.356 ± 0.041 | 6.3 ± 0.035 |
5.689 ± 0.036 | 6.992 ± 0.027 |
1.363 ± 0.018 | 2.86 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
