
Phytophthora megakarya
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Peronosporales; Peronosporaceae; Phytophthora
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
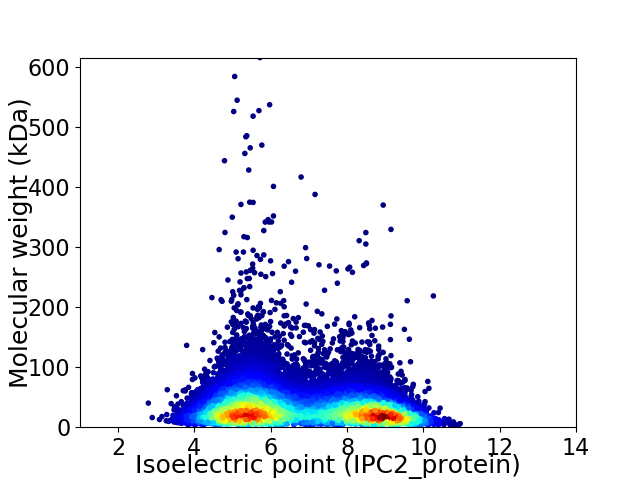
Virtual 2D-PAGE plot for 34747 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A225WHI1|A0A225WHI1_9STRA Exocyst subunit Exo70 family protein OS=Phytophthora megakarya OX=4795 GN=PHMEG_0009139 PE=3 SV=1
MM1 pKa = 7.97DD2 pKa = 5.42GSTTYY7 pKa = 10.37CYY9 pKa = 10.67YY10 pKa = 10.22ISEE13 pKa = 4.98KK14 pKa = 10.55YY15 pKa = 10.25SQEE18 pKa = 3.91QPVEE22 pKa = 4.17DD23 pKa = 4.79DD24 pKa = 3.97VLVMDD29 pKa = 5.24QGTKK33 pKa = 10.33CPITIRR39 pKa = 11.84VDD41 pKa = 3.02IADD44 pKa = 4.06EE45 pKa = 4.31LEE47 pKa = 4.24KK48 pKa = 11.09NALIPLRR55 pKa = 11.84YY56 pKa = 7.58TATLDD61 pKa = 3.63TSTADD66 pKa = 3.61ANFEE70 pKa = 4.09FPDD73 pKa = 5.0PIVKK77 pKa = 9.97VPVPDD82 pKa = 3.63YY83 pKa = 11.63LMSKK87 pKa = 8.83TNASTEE93 pKa = 4.28SSSSSDD99 pKa = 2.91TSFDD103 pKa = 3.39VAVANVVMCDD113 pKa = 3.01WRR115 pKa = 11.84SCNLFTTAGEE125 pKa = 4.18STTYY129 pKa = 10.47YY130 pKa = 10.61SSSNNPNDD138 pKa = 3.31FSDD141 pKa = 3.57NVAVFGSEE149 pKa = 3.76EE150 pKa = 4.0LVIPKK155 pKa = 9.91DD156 pKa = 3.66GKK158 pKa = 8.09YY159 pKa = 8.58TGYY162 pKa = 10.17VHH164 pKa = 7.06LVVNIGGNSRR174 pKa = 11.84ADD176 pKa = 3.23FVTFFPVQIGDD187 pKa = 3.76VAGTTPSSITTDD199 pKa = 3.1GTTTYY204 pKa = 10.06CWTSSDD210 pKa = 3.42VSVFDD215 pKa = 4.5PSVSGDD221 pKa = 3.22LTIRR225 pKa = 11.84TGEE228 pKa = 4.12YY229 pKa = 10.38CPGAMSMSLSSTDD242 pKa = 3.34VYY244 pKa = 11.46VGDD247 pKa = 5.03SIDD250 pKa = 3.76IAWMLDD256 pKa = 3.37MTDD259 pKa = 3.71SSTDD263 pKa = 3.32DD264 pKa = 3.24STLIAEE270 pKa = 4.67VSTTDD275 pKa = 4.45AIKK278 pKa = 10.58DD279 pKa = 3.7PRR281 pKa = 11.84NDD283 pKa = 2.98IYY285 pKa = 11.51SVVPVSVFSGCQRR298 pKa = 11.84NLAGANCSTYY308 pKa = 10.58TGDD311 pKa = 3.6EE312 pKa = 4.18SSTFAIAEE320 pKa = 4.01YY321 pKa = 10.87DD322 pKa = 3.74DD323 pKa = 4.28MNFTSDD329 pKa = 3.96AISYY333 pKa = 6.83STNYY337 pKa = 9.03TFEE340 pKa = 4.32NSGQYY345 pKa = 10.51TMFGRR350 pKa = 11.84VAMVTVDD357 pKa = 3.5GEE359 pKa = 4.48RR360 pKa = 11.84IDD362 pKa = 3.55MAIYY366 pKa = 10.34SSVTVSVDD374 pKa = 3.08GGSSGSNLFLYY385 pKa = 10.24IGIGIGAAILLGGLLFCYY403 pKa = 8.84MKK405 pKa = 10.51KK406 pKa = 10.15RR407 pKa = 11.84RR408 pKa = 11.84NNVNTKK414 pKa = 10.02DD415 pKa = 3.32IPFRR419 pKa = 11.84QPVGVMDD426 pKa = 4.65MDD428 pKa = 4.26RR429 pKa = 11.84SSDD432 pKa = 3.44FTMASSQFLTHH443 pKa = 6.24KK444 pKa = 9.71TPAQLHH450 pKa = 5.7GLDD453 pKa = 3.39MSGDD457 pKa = 3.58YY458 pKa = 10.47SANNTAPSYY467 pKa = 10.53LAINAGDD474 pKa = 3.45TSIFDD479 pKa = 3.74KK480 pKa = 10.87TISAEE485 pKa = 3.98PGQMAEE491 pKa = 4.29SVEE494 pKa = 4.29SSEE497 pKa = 4.89SFSLNPFEE505 pKa = 4.5RR506 pKa = 11.84ASFAGLSDD514 pKa = 4.24DD515 pKa = 4.13VASSRR520 pKa = 11.84PSDD523 pKa = 3.23MTKK526 pKa = 10.42FSFQSGYY533 pKa = 11.02DD534 pKa = 3.98DD535 pKa = 4.17GSEE538 pKa = 4.22WEE540 pKa = 5.08FDD542 pKa = 3.56TQNIGGDD549 pKa = 3.41RR550 pKa = 11.84GSDD553 pKa = 3.36FSDD556 pKa = 4.04FASNPGTSLPTTGTYY571 pKa = 10.72EE572 pKa = 4.14MGRR575 pKa = 11.84GMPILEE581 pKa = 4.4EE582 pKa = 4.6DD583 pKa = 4.75DD584 pKa = 5.27RR585 pKa = 11.84DD586 pKa = 3.61TDD588 pKa = 3.73TYY590 pKa = 11.41NPQHH594 pKa = 6.5QSTTTVDD601 pKa = 3.44PRR603 pKa = 11.84STTSSGWTVQQ613 pKa = 3.03
MM1 pKa = 7.97DD2 pKa = 5.42GSTTYY7 pKa = 10.37CYY9 pKa = 10.67YY10 pKa = 10.22ISEE13 pKa = 4.98KK14 pKa = 10.55YY15 pKa = 10.25SQEE18 pKa = 3.91QPVEE22 pKa = 4.17DD23 pKa = 4.79DD24 pKa = 3.97VLVMDD29 pKa = 5.24QGTKK33 pKa = 10.33CPITIRR39 pKa = 11.84VDD41 pKa = 3.02IADD44 pKa = 4.06EE45 pKa = 4.31LEE47 pKa = 4.24KK48 pKa = 11.09NALIPLRR55 pKa = 11.84YY56 pKa = 7.58TATLDD61 pKa = 3.63TSTADD66 pKa = 3.61ANFEE70 pKa = 4.09FPDD73 pKa = 5.0PIVKK77 pKa = 9.97VPVPDD82 pKa = 3.63YY83 pKa = 11.63LMSKK87 pKa = 8.83TNASTEE93 pKa = 4.28SSSSSDD99 pKa = 2.91TSFDD103 pKa = 3.39VAVANVVMCDD113 pKa = 3.01WRR115 pKa = 11.84SCNLFTTAGEE125 pKa = 4.18STTYY129 pKa = 10.47YY130 pKa = 10.61SSSNNPNDD138 pKa = 3.31FSDD141 pKa = 3.57NVAVFGSEE149 pKa = 3.76EE150 pKa = 4.0LVIPKK155 pKa = 9.91DD156 pKa = 3.66GKK158 pKa = 8.09YY159 pKa = 8.58TGYY162 pKa = 10.17VHH164 pKa = 7.06LVVNIGGNSRR174 pKa = 11.84ADD176 pKa = 3.23FVTFFPVQIGDD187 pKa = 3.76VAGTTPSSITTDD199 pKa = 3.1GTTTYY204 pKa = 10.06CWTSSDD210 pKa = 3.42VSVFDD215 pKa = 4.5PSVSGDD221 pKa = 3.22LTIRR225 pKa = 11.84TGEE228 pKa = 4.12YY229 pKa = 10.38CPGAMSMSLSSTDD242 pKa = 3.34VYY244 pKa = 11.46VGDD247 pKa = 5.03SIDD250 pKa = 3.76IAWMLDD256 pKa = 3.37MTDD259 pKa = 3.71SSTDD263 pKa = 3.32DD264 pKa = 3.24STLIAEE270 pKa = 4.67VSTTDD275 pKa = 4.45AIKK278 pKa = 10.58DD279 pKa = 3.7PRR281 pKa = 11.84NDD283 pKa = 2.98IYY285 pKa = 11.51SVVPVSVFSGCQRR298 pKa = 11.84NLAGANCSTYY308 pKa = 10.58TGDD311 pKa = 3.6EE312 pKa = 4.18SSTFAIAEE320 pKa = 4.01YY321 pKa = 10.87DD322 pKa = 3.74DD323 pKa = 4.28MNFTSDD329 pKa = 3.96AISYY333 pKa = 6.83STNYY337 pKa = 9.03TFEE340 pKa = 4.32NSGQYY345 pKa = 10.51TMFGRR350 pKa = 11.84VAMVTVDD357 pKa = 3.5GEE359 pKa = 4.48RR360 pKa = 11.84IDD362 pKa = 3.55MAIYY366 pKa = 10.34SSVTVSVDD374 pKa = 3.08GGSSGSNLFLYY385 pKa = 10.24IGIGIGAAILLGGLLFCYY403 pKa = 8.84MKK405 pKa = 10.51KK406 pKa = 10.15RR407 pKa = 11.84RR408 pKa = 11.84NNVNTKK414 pKa = 10.02DD415 pKa = 3.32IPFRR419 pKa = 11.84QPVGVMDD426 pKa = 4.65MDD428 pKa = 4.26RR429 pKa = 11.84SSDD432 pKa = 3.44FTMASSQFLTHH443 pKa = 6.24KK444 pKa = 9.71TPAQLHH450 pKa = 5.7GLDD453 pKa = 3.39MSGDD457 pKa = 3.58YY458 pKa = 10.47SANNTAPSYY467 pKa = 10.53LAINAGDD474 pKa = 3.45TSIFDD479 pKa = 3.74KK480 pKa = 10.87TISAEE485 pKa = 3.98PGQMAEE491 pKa = 4.29SVEE494 pKa = 4.29SSEE497 pKa = 4.89SFSLNPFEE505 pKa = 4.5RR506 pKa = 11.84ASFAGLSDD514 pKa = 4.24DD515 pKa = 4.13VASSRR520 pKa = 11.84PSDD523 pKa = 3.23MTKK526 pKa = 10.42FSFQSGYY533 pKa = 11.02DD534 pKa = 3.98DD535 pKa = 4.17GSEE538 pKa = 4.22WEE540 pKa = 5.08FDD542 pKa = 3.56TQNIGGDD549 pKa = 3.41RR550 pKa = 11.84GSDD553 pKa = 3.36FSDD556 pKa = 4.04FASNPGTSLPTTGTYY571 pKa = 10.72EE572 pKa = 4.14MGRR575 pKa = 11.84GMPILEE581 pKa = 4.4EE582 pKa = 4.6DD583 pKa = 4.75DD584 pKa = 5.27RR585 pKa = 11.84DD586 pKa = 3.61TDD588 pKa = 3.73TYY590 pKa = 11.41NPQHH594 pKa = 6.5QSTTTVDD601 pKa = 3.44PRR603 pKa = 11.84STTSSGWTVQQ613 pKa = 3.03
Molecular weight: 66.27 kDa
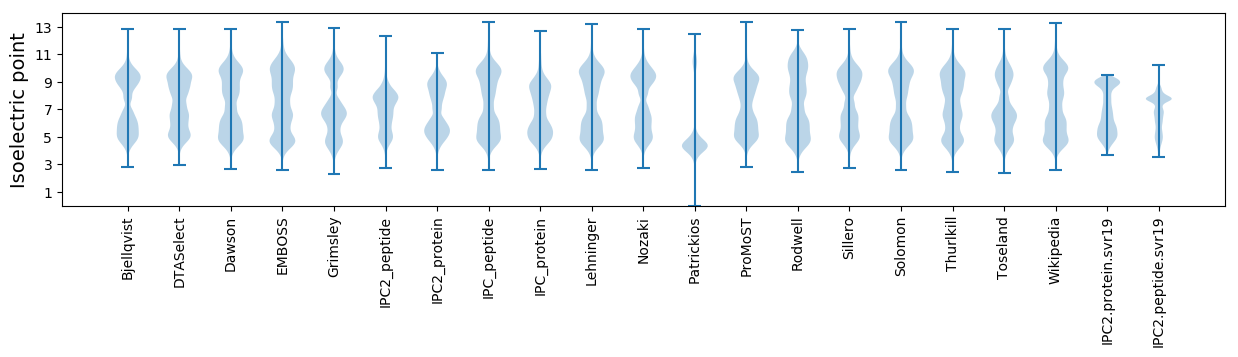
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A225UGQ9|A0A225UGQ9_9STRA Reverse transcriptase OS=Phytophthora megakarya OX=4795 GN=PHMEG_00038855 PE=4 SV=1
MM1 pKa = 7.61LISRR5 pKa = 11.84LHH7 pKa = 5.39GAPRR11 pKa = 11.84LLLLLRR17 pKa = 11.84SLKK20 pKa = 10.17VRR22 pKa = 11.84LPRR25 pKa = 11.84LPKK28 pKa = 9.83RR29 pKa = 11.84ARR31 pKa = 11.84STLSALLQLLLRR43 pKa = 11.84QQ44 pKa = 3.83
MM1 pKa = 7.61LISRR5 pKa = 11.84LHH7 pKa = 5.39GAPRR11 pKa = 11.84LLLLLRR17 pKa = 11.84SLKK20 pKa = 10.17VRR22 pKa = 11.84LPRR25 pKa = 11.84LPKK28 pKa = 9.83RR29 pKa = 11.84ARR31 pKa = 11.84STLSALLQLLLRR43 pKa = 11.84QQ44 pKa = 3.83
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10966191 |
9 |
7597 |
315.6 |
35.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.082 ± 0.013 | 1.667 ± 0.007 |
5.894 ± 0.011 | 6.482 ± 0.013 |
3.719 ± 0.008 | 5.844 ± 0.014 |
2.469 ± 0.006 | 4.466 ± 0.01 |
5.359 ± 0.015 | 9.196 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.552 ± 0.007 | 3.753 ± 0.008 |
4.734 ± 0.011 | 4.078 ± 0.01 |
6.606 ± 0.013 | 7.73 ± 0.017 |
6.044 ± 0.012 | 7.063 ± 0.012 |
1.427 ± 0.005 | 2.788 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
