
Raspberry vein chlorosis virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus; Raspberry vein chlorosis cytorhabdovirus
Average proteome isoelectric point is 7.47
Get precalculated fractions of proteins
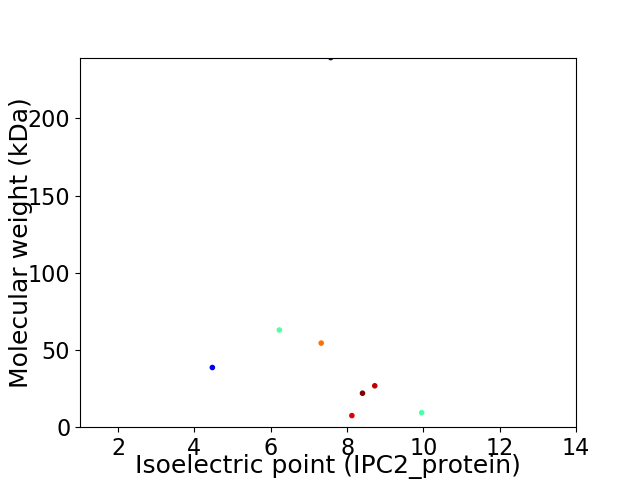
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A482P9Z3|A0A482P9Z3_9RHAB P6 protein OS=Raspberry vein chlorosis virus OX=758677 GN=P6 PE=4 SV=1
MM1 pKa = 8.1DD2 pKa = 6.21NIDD5 pKa = 5.42FDD7 pKa = 4.82ALPNPILDD15 pKa = 3.66VAMSEE20 pKa = 4.0IDD22 pKa = 3.52QEE24 pKa = 4.57RR25 pKa = 11.84NTDD28 pKa = 3.6DD29 pKa = 4.14LVAGKK34 pKa = 10.1SRR36 pKa = 11.84EE37 pKa = 4.17SNMGRR42 pKa = 11.84IEE44 pKa = 4.11EE45 pKa = 4.31SGQEE49 pKa = 4.1VAEE52 pKa = 4.24KK53 pKa = 10.21QSKK56 pKa = 10.44LEE58 pKa = 3.82SDD60 pKa = 3.58GAYY63 pKa = 10.14QEE65 pKa = 5.54DD66 pKa = 3.8IPYY69 pKa = 10.61DD70 pKa = 3.87RR71 pKa = 11.84NDD73 pKa = 3.29IDD75 pKa = 3.77TALTDD80 pKa = 4.13LQHH83 pKa = 6.88LCDD86 pKa = 4.26AMGVNYY92 pKa = 7.83TIPMEE97 pKa = 4.42NQVKK101 pKa = 9.32MLFRR105 pKa = 11.84DD106 pKa = 3.61EE107 pKa = 4.42EE108 pKa = 4.19VCYY111 pKa = 9.53THH113 pKa = 7.37LIWYY117 pKa = 8.99LRR119 pKa = 11.84GIINANQTQIIPTITGAISDD139 pKa = 3.99MKK141 pKa = 10.57MEE143 pKa = 4.15TRR145 pKa = 11.84QLQASSNKK153 pKa = 9.63LSKK156 pKa = 9.48EE157 pKa = 3.81TANIEE162 pKa = 4.2KK163 pKa = 9.68TSKK166 pKa = 10.55SLYY169 pKa = 10.4DD170 pKa = 3.85EE171 pKa = 3.86IRR173 pKa = 11.84AIKK176 pKa = 10.31EE177 pKa = 3.7DD178 pKa = 3.42MKK180 pKa = 11.12EE181 pKa = 3.98SFRR184 pKa = 11.84ASMKK188 pKa = 10.71LFMEE192 pKa = 5.06EE193 pKa = 4.02VQNEE197 pKa = 4.36KK198 pKa = 8.97PTQEE202 pKa = 4.08PKK204 pKa = 10.57QPLINPSKK212 pKa = 10.48IADD215 pKa = 3.87LKK217 pKa = 10.55AVKK220 pKa = 10.42DD221 pKa = 3.91LVLNSYY227 pKa = 10.6GNAGEE232 pKa = 4.8AMIPNKK238 pKa = 10.39ADD240 pKa = 3.09IPEE243 pKa = 4.36PTHH246 pKa = 4.8TNKK249 pKa = 10.01VIDD252 pKa = 4.52EE253 pKa = 4.15YY254 pKa = 11.39HH255 pKa = 5.98KK256 pKa = 10.58EE257 pKa = 3.93KK258 pKa = 10.57RR259 pKa = 11.84DD260 pKa = 3.57ALIKK264 pKa = 10.88YY265 pKa = 9.02GVDD268 pKa = 3.12PALVKK273 pKa = 10.58SYY275 pKa = 10.46EE276 pKa = 4.12DD277 pKa = 3.56SVIDD281 pKa = 4.84AMYY284 pKa = 10.0PDD286 pKa = 5.43DD287 pKa = 4.37VHH289 pKa = 7.13AQLKK293 pKa = 10.28AIRR296 pKa = 11.84LNEE299 pKa = 4.52GIKK302 pKa = 10.02RR303 pKa = 11.84QIRR306 pKa = 11.84KK307 pKa = 9.5SLEE310 pKa = 3.72EE311 pKa = 3.91RR312 pKa = 11.84LEE314 pKa = 4.62DD315 pKa = 3.85YY316 pKa = 11.06LDD318 pKa = 4.27EE319 pKa = 4.57EE320 pKa = 5.65DD321 pKa = 4.78EE322 pKa = 4.74DD323 pKa = 5.17EE324 pKa = 4.3EE325 pKa = 4.31MSNGSIQGDD334 pKa = 3.89SYY336 pKa = 12.07ASTEE340 pKa = 3.86
MM1 pKa = 8.1DD2 pKa = 6.21NIDD5 pKa = 5.42FDD7 pKa = 4.82ALPNPILDD15 pKa = 3.66VAMSEE20 pKa = 4.0IDD22 pKa = 3.52QEE24 pKa = 4.57RR25 pKa = 11.84NTDD28 pKa = 3.6DD29 pKa = 4.14LVAGKK34 pKa = 10.1SRR36 pKa = 11.84EE37 pKa = 4.17SNMGRR42 pKa = 11.84IEE44 pKa = 4.11EE45 pKa = 4.31SGQEE49 pKa = 4.1VAEE52 pKa = 4.24KK53 pKa = 10.21QSKK56 pKa = 10.44LEE58 pKa = 3.82SDD60 pKa = 3.58GAYY63 pKa = 10.14QEE65 pKa = 5.54DD66 pKa = 3.8IPYY69 pKa = 10.61DD70 pKa = 3.87RR71 pKa = 11.84NDD73 pKa = 3.29IDD75 pKa = 3.77TALTDD80 pKa = 4.13LQHH83 pKa = 6.88LCDD86 pKa = 4.26AMGVNYY92 pKa = 7.83TIPMEE97 pKa = 4.42NQVKK101 pKa = 9.32MLFRR105 pKa = 11.84DD106 pKa = 3.61EE107 pKa = 4.42EE108 pKa = 4.19VCYY111 pKa = 9.53THH113 pKa = 7.37LIWYY117 pKa = 8.99LRR119 pKa = 11.84GIINANQTQIIPTITGAISDD139 pKa = 3.99MKK141 pKa = 10.57MEE143 pKa = 4.15TRR145 pKa = 11.84QLQASSNKK153 pKa = 9.63LSKK156 pKa = 9.48EE157 pKa = 3.81TANIEE162 pKa = 4.2KK163 pKa = 9.68TSKK166 pKa = 10.55SLYY169 pKa = 10.4DD170 pKa = 3.85EE171 pKa = 3.86IRR173 pKa = 11.84AIKK176 pKa = 10.31EE177 pKa = 3.7DD178 pKa = 3.42MKK180 pKa = 11.12EE181 pKa = 3.98SFRR184 pKa = 11.84ASMKK188 pKa = 10.71LFMEE192 pKa = 5.06EE193 pKa = 4.02VQNEE197 pKa = 4.36KK198 pKa = 8.97PTQEE202 pKa = 4.08PKK204 pKa = 10.57QPLINPSKK212 pKa = 10.48IADD215 pKa = 3.87LKK217 pKa = 10.55AVKK220 pKa = 10.42DD221 pKa = 3.91LVLNSYY227 pKa = 10.6GNAGEE232 pKa = 4.8AMIPNKK238 pKa = 10.39ADD240 pKa = 3.09IPEE243 pKa = 4.36PTHH246 pKa = 4.8TNKK249 pKa = 10.01VIDD252 pKa = 4.52EE253 pKa = 4.15YY254 pKa = 11.39HH255 pKa = 5.98KK256 pKa = 10.58EE257 pKa = 3.93KK258 pKa = 10.57RR259 pKa = 11.84DD260 pKa = 3.57ALIKK264 pKa = 10.88YY265 pKa = 9.02GVDD268 pKa = 3.12PALVKK273 pKa = 10.58SYY275 pKa = 10.46EE276 pKa = 4.12DD277 pKa = 3.56SVIDD281 pKa = 4.84AMYY284 pKa = 10.0PDD286 pKa = 5.43DD287 pKa = 4.37VHH289 pKa = 7.13AQLKK293 pKa = 10.28AIRR296 pKa = 11.84LNEE299 pKa = 4.52GIKK302 pKa = 10.02RR303 pKa = 11.84QIRR306 pKa = 11.84KK307 pKa = 9.5SLEE310 pKa = 3.72EE311 pKa = 3.91RR312 pKa = 11.84LEE314 pKa = 4.62DD315 pKa = 3.85YY316 pKa = 11.06LDD318 pKa = 4.27EE319 pKa = 4.57EE320 pKa = 5.65DD321 pKa = 4.78EE322 pKa = 4.74DD323 pKa = 5.17EE324 pKa = 4.3EE325 pKa = 4.31MSNGSIQGDD334 pKa = 3.89SYY336 pKa = 12.07ASTEE340 pKa = 3.86
Molecular weight: 38.67 kDa
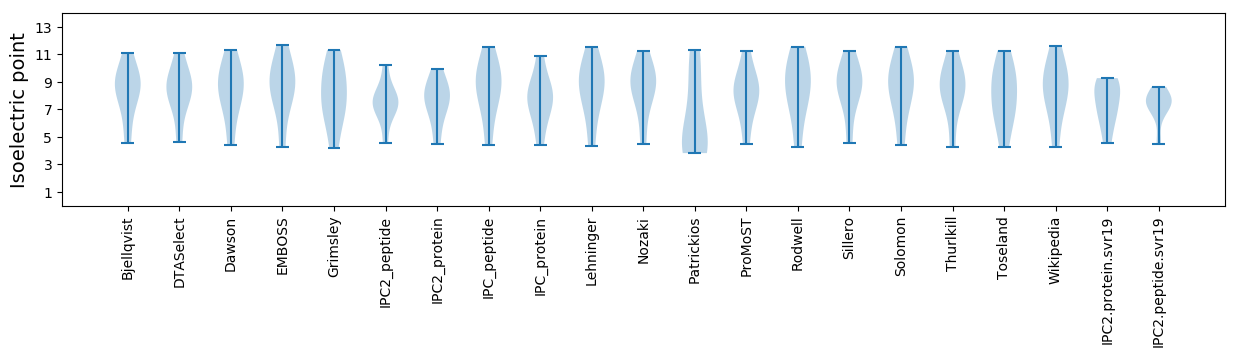
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A482PGG3|A0A482PGG3_9RHAB Movement protein OS=Raspberry vein chlorosis virus OX=758677 GN=P3 PE=4 SV=1
MM1 pKa = 7.11FVDD4 pKa = 4.51VDD6 pKa = 3.81PEE8 pKa = 4.24LLFKK12 pKa = 10.95LFSRR16 pKa = 11.84FISPNFLRR24 pKa = 11.84WTILILTLYY33 pKa = 8.49QTLSLMWQCLKK44 pKa = 10.65LIKK47 pKa = 10.47SVIQMILWLAKK58 pKa = 9.97AGRR61 pKa = 11.84AIWAGLKK68 pKa = 10.43NLGKK72 pKa = 10.36RR73 pKa = 11.84LRR75 pKa = 11.84RR76 pKa = 11.84SKK78 pKa = 11.18ASS80 pKa = 3.16
MM1 pKa = 7.11FVDD4 pKa = 4.51VDD6 pKa = 3.81PEE8 pKa = 4.24LLFKK12 pKa = 10.95LFSRR16 pKa = 11.84FISPNFLRR24 pKa = 11.84WTILILTLYY33 pKa = 8.49QTLSLMWQCLKK44 pKa = 10.65LIKK47 pKa = 10.47SVIQMILWLAKK58 pKa = 9.97AGRR61 pKa = 11.84AIWAGLKK68 pKa = 10.43NLGKK72 pKa = 10.36RR73 pKa = 11.84LRR75 pKa = 11.84RR76 pKa = 11.84SKK78 pKa = 11.18ASS80 pKa = 3.16
Molecular weight: 9.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4072 |
65 |
2092 |
509.0 |
57.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.231 ± 1.318 | 1.817 ± 0.354 |
6.238 ± 0.656 | 5.968 ± 0.744 |
2.947 ± 0.332 | 5.869 ± 0.628 |
2.112 ± 0.329 | 7.22 ± 0.458 |
6.68 ± 0.338 | 9.234 ± 0.757 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.88 ± 0.259 | 4.47 ± 0.34 |
4.052 ± 0.232 | 2.849 ± 0.464 |
5.82 ± 0.381 | 8.374 ± 0.435 |
6.164 ± 0.393 | 5.918 ± 0.385 |
1.523 ± 0.259 | 3.635 ± 0.341 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
