
Infectious hematopoietic necrosis virus (strain WRAC) (IHNV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Gammarhabdovirinae; Novirhabdovirus; Salmonid novirhabdovirus; Infectious hematopoietic necrosis virus
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
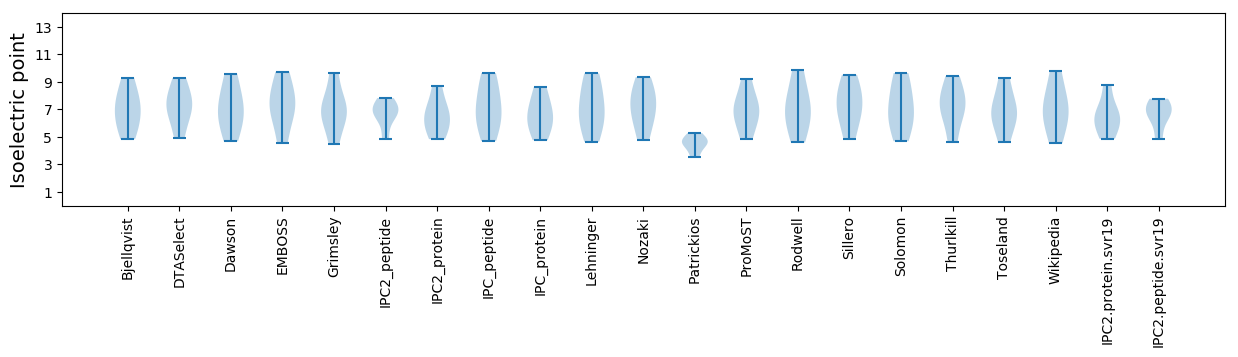
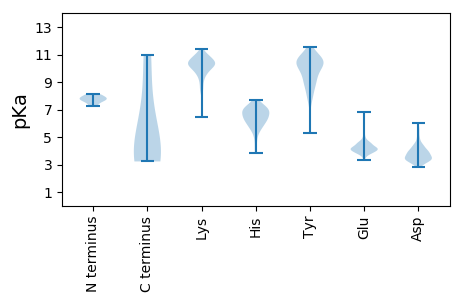
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q82681|PHOSP_IHNVW Phosphoprotein OS=Infectious hematopoietic necrosis virus (strain WRAC) OX=429314 GN=P PE=3 SV=1
MM1 pKa = 7.27TSALRR6 pKa = 11.84EE7 pKa = 4.31TFTGLRR13 pKa = 11.84DD14 pKa = 3.4IKK16 pKa = 11.01GGVLEE21 pKa = 5.08DD22 pKa = 4.37PEE24 pKa = 4.5TEE26 pKa = 4.17YY27 pKa = 11.12RR28 pKa = 11.84PSTITLPLFFSKK40 pKa = 9.93TDD42 pKa = 3.96LDD44 pKa = 3.96LEE46 pKa = 4.45MIKK49 pKa = 10.41RR50 pKa = 11.84AVSQVGGEE58 pKa = 4.35GTRR61 pKa = 11.84KK62 pKa = 9.93ALSLLCAFVIAEE74 pKa = 4.46TVPSEE79 pKa = 4.18AGTVAEE85 pKa = 4.58LLEE88 pKa = 4.18ALGFVLEE95 pKa = 4.52SLDD98 pKa = 3.49TGAPPDD104 pKa = 3.76ATFADD109 pKa = 4.49PNNKK113 pKa = 9.42LAEE116 pKa = 4.54TIVKK120 pKa = 10.04EE121 pKa = 4.02NVLEE125 pKa = 4.28VVTGLLFTCALLTKK139 pKa = 10.44YY140 pKa = 10.51DD141 pKa = 3.65VDD143 pKa = 5.43KK144 pKa = 10.91MATYY148 pKa = 9.75CQNKK152 pKa = 9.71LEE154 pKa = 4.45RR155 pKa = 11.84LATSQGIGEE164 pKa = 4.45LVNFNANRR172 pKa = 11.84GVLAKK177 pKa = 9.98IGAVLRR183 pKa = 11.84PGQKK187 pKa = 8.37LTKK190 pKa = 9.73AIYY193 pKa = 9.93GIILINLSDD202 pKa = 3.73PAIAARR208 pKa = 11.84AKK210 pKa = 10.03ALCAMRR216 pKa = 11.84LSGTGMTMVGLFNQAAKK233 pKa = 10.88NLGALPADD241 pKa = 4.15LLEE244 pKa = 4.56DD245 pKa = 3.65LCMKK249 pKa = 10.5SVVEE253 pKa = 3.85SARR256 pKa = 11.84RR257 pKa = 11.84IVRR260 pKa = 11.84LMRR263 pKa = 11.84IVAEE267 pKa = 4.31APGVAAKK274 pKa = 10.28YY275 pKa = 9.64GVMMSRR281 pKa = 11.84MLGEE285 pKa = 5.19GYY287 pKa = 10.46FKK289 pKa = 10.86AYY291 pKa = 10.1GINEE295 pKa = 4.17NARR298 pKa = 11.84ITCILMNINDD308 pKa = 4.56RR309 pKa = 11.84YY310 pKa = 11.36DD311 pKa = 3.95DD312 pKa = 3.92GTSRR316 pKa = 11.84GLTGIKK322 pKa = 10.33VSDD325 pKa = 4.27PFRR328 pKa = 11.84KK329 pKa = 9.43LARR332 pKa = 11.84EE333 pKa = 3.85IARR336 pKa = 11.84LLVLKK341 pKa = 10.68YY342 pKa = 10.73DD343 pKa = 4.17GDD345 pKa = 4.03GSTGEE350 pKa = 4.37GASEE354 pKa = 4.73LIRR357 pKa = 11.84RR358 pKa = 11.84AEE360 pKa = 3.99MASRR364 pKa = 11.84GPDD367 pKa = 2.89MGEE370 pKa = 3.74EE371 pKa = 4.11EE372 pKa = 4.82EE373 pKa = 4.79EE374 pKa = 4.85DD375 pKa = 4.39EE376 pKa = 4.91EE377 pKa = 6.79DD378 pKa = 5.06DD379 pKa = 5.57DD380 pKa = 5.65SSEE383 pKa = 4.67PGDD386 pKa = 3.5SDD388 pKa = 4.46SFHH391 pKa = 6.93
MM1 pKa = 7.27TSALRR6 pKa = 11.84EE7 pKa = 4.31TFTGLRR13 pKa = 11.84DD14 pKa = 3.4IKK16 pKa = 11.01GGVLEE21 pKa = 5.08DD22 pKa = 4.37PEE24 pKa = 4.5TEE26 pKa = 4.17YY27 pKa = 11.12RR28 pKa = 11.84PSTITLPLFFSKK40 pKa = 9.93TDD42 pKa = 3.96LDD44 pKa = 3.96LEE46 pKa = 4.45MIKK49 pKa = 10.41RR50 pKa = 11.84AVSQVGGEE58 pKa = 4.35GTRR61 pKa = 11.84KK62 pKa = 9.93ALSLLCAFVIAEE74 pKa = 4.46TVPSEE79 pKa = 4.18AGTVAEE85 pKa = 4.58LLEE88 pKa = 4.18ALGFVLEE95 pKa = 4.52SLDD98 pKa = 3.49TGAPPDD104 pKa = 3.76ATFADD109 pKa = 4.49PNNKK113 pKa = 9.42LAEE116 pKa = 4.54TIVKK120 pKa = 10.04EE121 pKa = 4.02NVLEE125 pKa = 4.28VVTGLLFTCALLTKK139 pKa = 10.44YY140 pKa = 10.51DD141 pKa = 3.65VDD143 pKa = 5.43KK144 pKa = 10.91MATYY148 pKa = 9.75CQNKK152 pKa = 9.71LEE154 pKa = 4.45RR155 pKa = 11.84LATSQGIGEE164 pKa = 4.45LVNFNANRR172 pKa = 11.84GVLAKK177 pKa = 9.98IGAVLRR183 pKa = 11.84PGQKK187 pKa = 8.37LTKK190 pKa = 9.73AIYY193 pKa = 9.93GIILINLSDD202 pKa = 3.73PAIAARR208 pKa = 11.84AKK210 pKa = 10.03ALCAMRR216 pKa = 11.84LSGTGMTMVGLFNQAAKK233 pKa = 10.88NLGALPADD241 pKa = 4.15LLEE244 pKa = 4.56DD245 pKa = 3.65LCMKK249 pKa = 10.5SVVEE253 pKa = 3.85SARR256 pKa = 11.84RR257 pKa = 11.84IVRR260 pKa = 11.84LMRR263 pKa = 11.84IVAEE267 pKa = 4.31APGVAAKK274 pKa = 10.28YY275 pKa = 9.64GVMMSRR281 pKa = 11.84MLGEE285 pKa = 5.19GYY287 pKa = 10.46FKK289 pKa = 10.86AYY291 pKa = 10.1GINEE295 pKa = 4.17NARR298 pKa = 11.84ITCILMNINDD308 pKa = 4.56RR309 pKa = 11.84YY310 pKa = 11.36DD311 pKa = 3.95DD312 pKa = 3.92GTSRR316 pKa = 11.84GLTGIKK322 pKa = 10.33VSDD325 pKa = 4.27PFRR328 pKa = 11.84KK329 pKa = 9.43LARR332 pKa = 11.84EE333 pKa = 3.85IARR336 pKa = 11.84LLVLKK341 pKa = 10.68YY342 pKa = 10.73DD343 pKa = 4.17GDD345 pKa = 4.03GSTGEE350 pKa = 4.37GASEE354 pKa = 4.73LIRR357 pKa = 11.84RR358 pKa = 11.84AEE360 pKa = 3.99MASRR364 pKa = 11.84GPDD367 pKa = 2.89MGEE370 pKa = 3.74EE371 pKa = 4.11EE372 pKa = 4.82EE373 pKa = 4.79EE374 pKa = 4.85DD375 pKa = 4.39EE376 pKa = 4.91EE377 pKa = 6.79DD378 pKa = 5.06DD379 pKa = 5.57DD380 pKa = 5.65SSEE383 pKa = 4.67PGDD386 pKa = 3.5SDD388 pKa = 4.46SFHH391 pKa = 6.93
Molecular weight: 42.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q82683|GLYCO_IHNVW Glycoprotein OS=Infectious hematopoietic necrosis virus (strain WRAC) OX=429314 GN=G PE=3 SV=1
MM1 pKa = 7.66SIFKK5 pKa = 10.15RR6 pKa = 11.84AKK8 pKa = 8.38KK9 pKa = 8.4TVLIPPPHH17 pKa = 6.69LLSGDD22 pKa = 3.82EE23 pKa = 4.03EE24 pKa = 4.63RR25 pKa = 11.84VTILSAEE32 pKa = 4.3GEE34 pKa = 4.21IKK36 pKa = 9.74VTGRR40 pKa = 11.84RR41 pKa = 11.84PTTLEE46 pKa = 3.5EE47 pKa = 4.2KK48 pKa = 9.99IYY50 pKa = 11.25YY51 pKa = 10.45SMNLAAAIVGGDD63 pKa = 3.31LHH65 pKa = 7.04PSFKK69 pKa = 11.13SMTYY73 pKa = 10.43LFQKK77 pKa = 10.35EE78 pKa = 4.28MEE80 pKa = 4.5FGSTQEE86 pKa = 3.99KK87 pKa = 10.7VNFGSRR93 pKa = 11.84KK94 pKa = 8.71PAPQTTYY101 pKa = 10.34QVTKK105 pKa = 10.32ARR107 pKa = 11.84EE108 pKa = 4.32VYY110 pKa = 10.51LQTQPLEE117 pKa = 4.38KK118 pKa = 9.74KK119 pKa = 10.35IPMQTYY125 pKa = 9.21SVSTEE130 pKa = 3.61GATITFTGRR139 pKa = 11.84FLFSSSHH146 pKa = 6.01VGCDD150 pKa = 3.73DD151 pKa = 4.41NRR153 pKa = 11.84TKK155 pKa = 10.94LAGLDD160 pKa = 3.59GFTTSNSYY168 pKa = 10.23QRR170 pKa = 11.84VKK172 pKa = 10.7DD173 pKa = 3.79YY174 pKa = 11.04YY175 pKa = 10.56AQEE178 pKa = 3.85TALALTFAAPEE189 pKa = 3.83KK190 pKa = 10.28RR191 pKa = 11.84GKK193 pKa = 10.02EE194 pKa = 3.92KK195 pKa = 10.97
MM1 pKa = 7.66SIFKK5 pKa = 10.15RR6 pKa = 11.84AKK8 pKa = 8.38KK9 pKa = 8.4TVLIPPPHH17 pKa = 6.69LLSGDD22 pKa = 3.82EE23 pKa = 4.03EE24 pKa = 4.63RR25 pKa = 11.84VTILSAEE32 pKa = 4.3GEE34 pKa = 4.21IKK36 pKa = 9.74VTGRR40 pKa = 11.84RR41 pKa = 11.84PTTLEE46 pKa = 3.5EE47 pKa = 4.2KK48 pKa = 9.99IYY50 pKa = 11.25YY51 pKa = 10.45SMNLAAAIVGGDD63 pKa = 3.31LHH65 pKa = 7.04PSFKK69 pKa = 11.13SMTYY73 pKa = 10.43LFQKK77 pKa = 10.35EE78 pKa = 4.28MEE80 pKa = 4.5FGSTQEE86 pKa = 3.99KK87 pKa = 10.7VNFGSRR93 pKa = 11.84KK94 pKa = 8.71PAPQTTYY101 pKa = 10.34QVTKK105 pKa = 10.32ARR107 pKa = 11.84EE108 pKa = 4.32VYY110 pKa = 10.51LQTQPLEE117 pKa = 4.38KK118 pKa = 9.74KK119 pKa = 10.35IPMQTYY125 pKa = 9.21SVSTEE130 pKa = 3.61GATITFTGRR139 pKa = 11.84FLFSSSHH146 pKa = 6.01VGCDD150 pKa = 3.73DD151 pKa = 4.41NRR153 pKa = 11.84TKK155 pKa = 10.94LAGLDD160 pKa = 3.59GFTTSNSYY168 pKa = 10.23QRR170 pKa = 11.84VKK172 pKa = 10.7DD173 pKa = 3.79YY174 pKa = 11.04YY175 pKa = 10.56AQEE178 pKa = 3.85TALALTFAAPEE189 pKa = 3.83KK190 pKa = 10.28RR191 pKa = 11.84GKK193 pKa = 10.02EE194 pKa = 3.92KK195 pKa = 10.97
Molecular weight: 21.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3421 |
111 |
1986 |
570.2 |
64.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.782 ± 0.737 | 1.432 ± 0.367 |
5.817 ± 0.423 | 7.279 ± 0.723 |
3.712 ± 0.216 | 6.109 ± 0.586 |
2.163 ± 0.4 | 6.665 ± 0.763 |
6.226 ± 0.562 | 10.932 ± 1.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.338 ± 0.303 | 3.537 ± 0.289 |
4.443 ± 0.338 | 3.391 ± 0.583 |
6.402 ± 0.591 | 7.395 ± 0.642 |
6.519 ± 0.66 | 4.969 ± 0.253 |
1.257 ± 0.395 | 2.631 ± 0.446 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
