
Spinybacked orbweaver circular virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses; Circularisvirus
Average proteome isoelectric point is 8.85
Get precalculated fractions of proteins
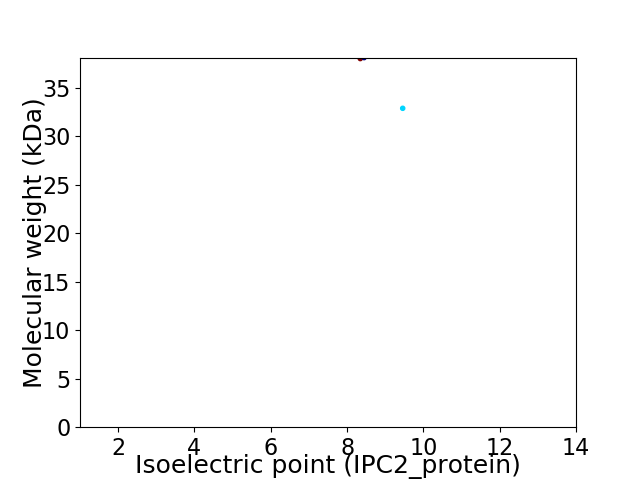
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPA1|A0A346BPA1_9VIRU Putative capsid protein OS=Spinybacked orbweaver circular virus 1 OX=2293305 PE=4 SV=1
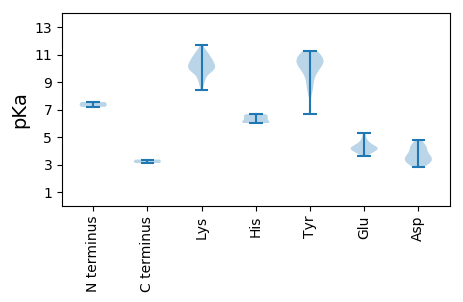
MM1 pKa = 7.19SRR3 pKa = 11.84CHH5 pKa = 6.04GWTFTVFALEE15 pKa = 4.24PEE17 pKa = 4.59PVFEE21 pKa = 5.09PNLHH25 pKa = 6.64EE26 pKa = 3.83YY27 pKa = 10.77LIFGRR32 pKa = 11.84EE33 pKa = 3.97TCPTTGRR40 pKa = 11.84SHH42 pKa = 6.18LQGYY46 pKa = 9.92IYY48 pKa = 10.4FKK50 pKa = 10.21EE51 pKa = 4.07RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84LPYY57 pKa = 9.69LKK59 pKa = 10.06KK60 pKa = 9.48WIPGAHH66 pKa = 6.02FEE68 pKa = 4.14AAKK71 pKa = 10.69GNPEE75 pKa = 3.9QNQKK79 pKa = 10.31YY80 pKa = 9.46CSKK83 pKa = 11.11DD84 pKa = 3.12GDD86 pKa = 3.73FKK88 pKa = 11.41EE89 pKa = 4.29YY90 pKa = 10.93GRR92 pKa = 11.84LPRR95 pKa = 11.84SKK97 pKa = 10.59SGGGAFKK104 pKa = 10.81DD105 pKa = 3.5VLLKK109 pKa = 10.97AEE111 pKa = 4.72SGNIADD117 pKa = 4.8IKK119 pKa = 10.8EE120 pKa = 4.23EE121 pKa = 4.21YY122 pKa = 9.4PGLFIRR128 pKa = 11.84YY129 pKa = 6.7KK130 pKa = 9.9TNILSSVKK138 pKa = 10.11FRR140 pKa = 11.84VEE142 pKa = 3.98EE143 pKa = 4.14LKK145 pKa = 10.97SSCGVWICGPPRR157 pKa = 11.84CGKK160 pKa = 10.26DD161 pKa = 3.24YY162 pKa = 11.18AVRR165 pKa = 11.84KK166 pKa = 9.71LNSVYY171 pKa = 10.7VKK173 pKa = 10.27PLNKK177 pKa = 9.2WWDD180 pKa = 3.72GYY182 pKa = 10.78NNEE185 pKa = 4.18NFVLLSDD192 pKa = 4.36VEE194 pKa = 4.45PPHH197 pKa = 6.66CSWLGFFLKK206 pKa = 10.08IWTDD210 pKa = 3.27RR211 pKa = 11.84YY212 pKa = 11.26AFVAEE217 pKa = 4.17IKK219 pKa = 10.54GGSVKK224 pKa = 9.84IRR226 pKa = 11.84PKK228 pKa = 10.61KK229 pKa = 10.45IFVTSNFTLEE239 pKa = 3.93EE240 pKa = 4.25CFSGKK245 pKa = 9.15VLEE248 pKa = 4.26ALRR251 pKa = 11.84TRR253 pKa = 11.84FNVYY257 pKa = 10.75NEE259 pKa = 3.81FDD261 pKa = 3.27GSFFARR267 pKa = 11.84QVDD270 pKa = 4.28SIKK273 pKa = 10.0TAVYY277 pKa = 10.45DD278 pKa = 3.85KK279 pKa = 11.04LLSLEE284 pKa = 4.79DD285 pKa = 3.69GLVQEE290 pKa = 5.28KK291 pKa = 10.69EE292 pKa = 4.17NVSPSVSKK300 pKa = 11.23AEE302 pKa = 4.6EE303 pKa = 3.89ILSSSEE309 pKa = 3.62EE310 pKa = 3.97FEE312 pKa = 4.95NVQKK316 pKa = 10.57PSKK319 pKa = 9.87KK320 pKa = 9.82KK321 pKa = 10.26KK322 pKa = 9.94RR323 pKa = 11.84LSKK326 pKa = 9.92TNSAQQ331 pKa = 3.11
MM1 pKa = 7.19SRR3 pKa = 11.84CHH5 pKa = 6.04GWTFTVFALEE15 pKa = 4.24PEE17 pKa = 4.59PVFEE21 pKa = 5.09PNLHH25 pKa = 6.64EE26 pKa = 3.83YY27 pKa = 10.77LIFGRR32 pKa = 11.84EE33 pKa = 3.97TCPTTGRR40 pKa = 11.84SHH42 pKa = 6.18LQGYY46 pKa = 9.92IYY48 pKa = 10.4FKK50 pKa = 10.21EE51 pKa = 4.07RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84LPYY57 pKa = 9.69LKK59 pKa = 10.06KK60 pKa = 9.48WIPGAHH66 pKa = 6.02FEE68 pKa = 4.14AAKK71 pKa = 10.69GNPEE75 pKa = 3.9QNQKK79 pKa = 10.31YY80 pKa = 9.46CSKK83 pKa = 11.11DD84 pKa = 3.12GDD86 pKa = 3.73FKK88 pKa = 11.41EE89 pKa = 4.29YY90 pKa = 10.93GRR92 pKa = 11.84LPRR95 pKa = 11.84SKK97 pKa = 10.59SGGGAFKK104 pKa = 10.81DD105 pKa = 3.5VLLKK109 pKa = 10.97AEE111 pKa = 4.72SGNIADD117 pKa = 4.8IKK119 pKa = 10.8EE120 pKa = 4.23EE121 pKa = 4.21YY122 pKa = 9.4PGLFIRR128 pKa = 11.84YY129 pKa = 6.7KK130 pKa = 9.9TNILSSVKK138 pKa = 10.11FRR140 pKa = 11.84VEE142 pKa = 3.98EE143 pKa = 4.14LKK145 pKa = 10.97SSCGVWICGPPRR157 pKa = 11.84CGKK160 pKa = 10.26DD161 pKa = 3.24YY162 pKa = 11.18AVRR165 pKa = 11.84KK166 pKa = 9.71LNSVYY171 pKa = 10.7VKK173 pKa = 10.27PLNKK177 pKa = 9.2WWDD180 pKa = 3.72GYY182 pKa = 10.78NNEE185 pKa = 4.18NFVLLSDD192 pKa = 4.36VEE194 pKa = 4.45PPHH197 pKa = 6.66CSWLGFFLKK206 pKa = 10.08IWTDD210 pKa = 3.27RR211 pKa = 11.84YY212 pKa = 11.26AFVAEE217 pKa = 4.17IKK219 pKa = 10.54GGSVKK224 pKa = 9.84IRR226 pKa = 11.84PKK228 pKa = 10.61KK229 pKa = 10.45IFVTSNFTLEE239 pKa = 3.93EE240 pKa = 4.25CFSGKK245 pKa = 9.15VLEE248 pKa = 4.26ALRR251 pKa = 11.84TRR253 pKa = 11.84FNVYY257 pKa = 10.75NEE259 pKa = 3.81FDD261 pKa = 3.27GSFFARR267 pKa = 11.84QVDD270 pKa = 4.28SIKK273 pKa = 10.0TAVYY277 pKa = 10.45DD278 pKa = 3.85KK279 pKa = 11.04LLSLEE284 pKa = 4.79DD285 pKa = 3.69GLVQEE290 pKa = 5.28KK291 pKa = 10.69EE292 pKa = 4.17NVSPSVSKK300 pKa = 11.23AEE302 pKa = 4.6EE303 pKa = 3.89ILSSSEE309 pKa = 3.62EE310 pKa = 3.97FEE312 pKa = 4.95NVQKK316 pKa = 10.57PSKK319 pKa = 9.87KK320 pKa = 9.82KK321 pKa = 10.26KK322 pKa = 9.94RR323 pKa = 11.84LSKK326 pKa = 9.92TNSAQQ331 pKa = 3.11
Molecular weight: 38.0 kDa
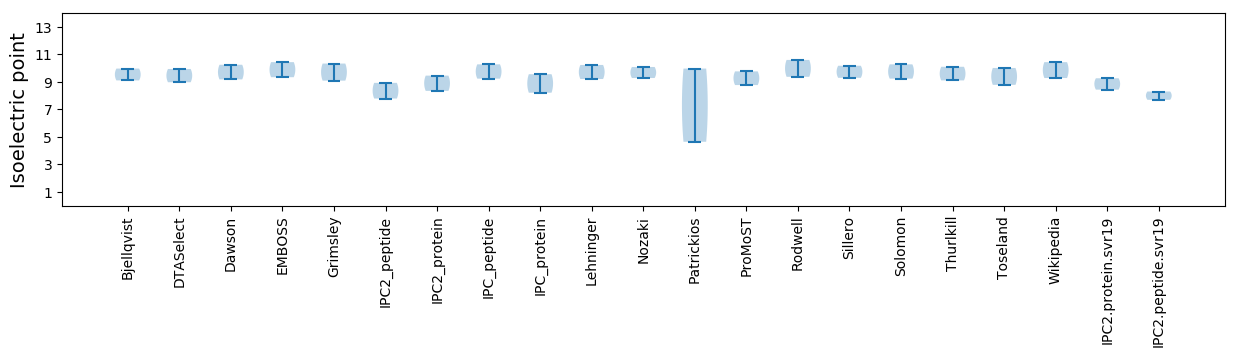
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPA1|A0A346BPA1_9VIRU Putative capsid protein OS=Spinybacked orbweaver circular virus 1 OX=2293305 PE=4 SV=1
MM1 pKa = 7.55AWFKK5 pKa = 10.36RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84MFRR11 pKa = 11.84RR12 pKa = 11.84LFRR15 pKa = 11.84KK16 pKa = 9.38RR17 pKa = 11.84KK18 pKa = 8.72KK19 pKa = 9.48FYY21 pKa = 9.89HH22 pKa = 6.03RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84NLRR28 pKa = 11.84MFRR31 pKa = 11.84NRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84NDD39 pKa = 2.84YY40 pKa = 10.53RR41 pKa = 11.84KK42 pKa = 8.42QTVLSDD48 pKa = 2.94ITYY51 pKa = 8.79TLDD54 pKa = 3.22VTHH57 pKa = 6.46EE58 pKa = 4.13QDD60 pKa = 4.76SKK62 pKa = 11.72SSTNNAEE69 pKa = 3.73MWGANCSLSALQTPEE84 pKa = 4.52FKK86 pKa = 10.07TLCKK90 pKa = 10.09DD91 pKa = 3.09WKK93 pKa = 9.01FCKK96 pKa = 9.97INSISYY102 pKa = 8.75ACTVGAIGYY111 pKa = 8.57NVYY114 pKa = 10.54EE115 pKa = 4.4PADD118 pKa = 3.56AKK120 pKa = 11.24AKK122 pKa = 10.05VQAQRR127 pKa = 11.84IDD129 pKa = 4.25LEE131 pKa = 4.42VNDD134 pKa = 4.25IVKK137 pKa = 9.78NQPFYY142 pKa = 10.67FIWDD146 pKa = 3.36ADD148 pKa = 3.6TRR150 pKa = 11.84LAEE153 pKa = 4.46SGNVNPNQLMSEE165 pKa = 4.31TSKK168 pKa = 10.58KK169 pKa = 9.95HH170 pKa = 6.42CIINGKK176 pKa = 9.82KK177 pKa = 9.84SAFFNLRR184 pKa = 11.84PPSILRR190 pKa = 11.84RR191 pKa = 11.84YY192 pKa = 8.24YY193 pKa = 10.52DD194 pKa = 3.18SKK196 pKa = 11.21VVYY199 pKa = 10.64NNRR202 pKa = 11.84NKK204 pKa = 8.97NTWGEE209 pKa = 4.14YY210 pKa = 10.01FDD212 pKa = 4.25NLSTISNYY220 pKa = 10.26RR221 pKa = 11.84MCNTFRR227 pKa = 11.84GTCGSYY233 pKa = 10.39FRR235 pKa = 11.84SLPIFVKK242 pKa = 10.62VDD244 pKa = 3.61DD245 pKa = 4.2TPVIYY250 pKa = 10.32KK251 pKa = 10.38CAMKK255 pKa = 10.72LILRR259 pKa = 11.84VKK261 pKa = 10.34CYY263 pKa = 10.8VNCTFKK269 pKa = 10.98GRR271 pKa = 11.84AYY273 pKa = 10.11EE274 pKa = 4.08YY275 pKa = 11.2NN276 pKa = 3.36
MM1 pKa = 7.55AWFKK5 pKa = 10.36RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84MFRR11 pKa = 11.84RR12 pKa = 11.84LFRR15 pKa = 11.84KK16 pKa = 9.38RR17 pKa = 11.84KK18 pKa = 8.72KK19 pKa = 9.48FYY21 pKa = 9.89HH22 pKa = 6.03RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84NLRR28 pKa = 11.84MFRR31 pKa = 11.84NRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84NDD39 pKa = 2.84YY40 pKa = 10.53RR41 pKa = 11.84KK42 pKa = 8.42QTVLSDD48 pKa = 2.94ITYY51 pKa = 8.79TLDD54 pKa = 3.22VTHH57 pKa = 6.46EE58 pKa = 4.13QDD60 pKa = 4.76SKK62 pKa = 11.72SSTNNAEE69 pKa = 3.73MWGANCSLSALQTPEE84 pKa = 4.52FKK86 pKa = 10.07TLCKK90 pKa = 10.09DD91 pKa = 3.09WKK93 pKa = 9.01FCKK96 pKa = 9.97INSISYY102 pKa = 8.75ACTVGAIGYY111 pKa = 8.57NVYY114 pKa = 10.54EE115 pKa = 4.4PADD118 pKa = 3.56AKK120 pKa = 11.24AKK122 pKa = 10.05VQAQRR127 pKa = 11.84IDD129 pKa = 4.25LEE131 pKa = 4.42VNDD134 pKa = 4.25IVKK137 pKa = 9.78NQPFYY142 pKa = 10.67FIWDD146 pKa = 3.36ADD148 pKa = 3.6TRR150 pKa = 11.84LAEE153 pKa = 4.46SGNVNPNQLMSEE165 pKa = 4.31TSKK168 pKa = 10.58KK169 pKa = 9.95HH170 pKa = 6.42CIINGKK176 pKa = 9.82KK177 pKa = 9.84SAFFNLRR184 pKa = 11.84PPSILRR190 pKa = 11.84RR191 pKa = 11.84YY192 pKa = 8.24YY193 pKa = 10.52DD194 pKa = 3.18SKK196 pKa = 11.21VVYY199 pKa = 10.64NNRR202 pKa = 11.84NKK204 pKa = 8.97NTWGEE209 pKa = 4.14YY210 pKa = 10.01FDD212 pKa = 4.25NLSTISNYY220 pKa = 10.26RR221 pKa = 11.84MCNTFRR227 pKa = 11.84GTCGSYY233 pKa = 10.39FRR235 pKa = 11.84SLPIFVKK242 pKa = 10.62VDD244 pKa = 3.61DD245 pKa = 4.2TPVIYY250 pKa = 10.32KK251 pKa = 10.38CAMKK255 pKa = 10.72LILRR259 pKa = 11.84VKK261 pKa = 10.34CYY263 pKa = 10.8VNCTFKK269 pKa = 10.98GRR271 pKa = 11.84AYY273 pKa = 10.11EE274 pKa = 4.08YY275 pKa = 11.2NN276 pKa = 3.36
Molecular weight: 32.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
607 |
276 |
331 |
303.5 |
35.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.942 ± 0.332 | 2.965 ± 0.443 |
4.283 ± 0.532 | 6.26 ± 2.021 |
6.26 ± 0.312 | 5.272 ± 1.355 |
1.318 ± 0.156 | 4.613 ± 0.31 |
9.555 ± 0.579 | 7.084 ± 0.867 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.318 ± 0.821 | 6.59 ± 1.419 |
4.119 ± 0.822 | 2.306 ± 0.155 |
7.743 ± 1.862 | 7.578 ± 0.712 |
4.613 ± 0.798 | 6.096 ± 0.445 |
1.977 ± 0.111 | 5.107 ± 0.709 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
