
Infectious pancreatic necrosis virus (strain Sp) (IPNV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Birnaviridae; Aquabirnavirus; Infectious pancreatic necrosis virus
Average proteome isoelectric point is 5.68
Get precalculated fractions of proteins
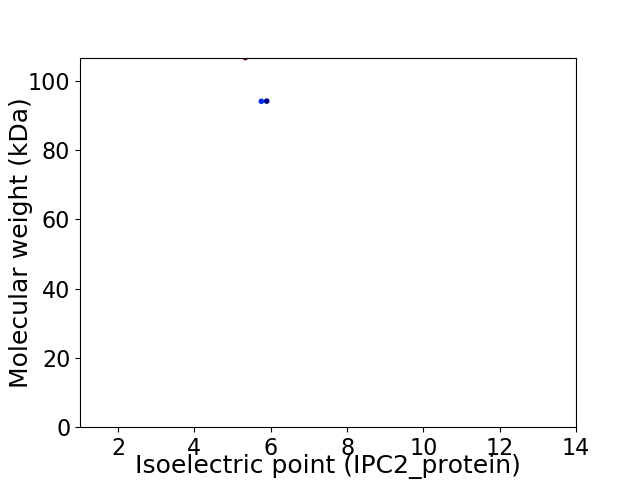
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q703G8|Q703G8_IPNVS Isoform of P22174 Protein VP1 OS=Infectious pancreatic necrosis virus (strain Sp) OX=11005 GN=VP1 PE=4 SV=1
MM1 pKa = 6.89NTNKK5 pKa = 8.97ATATYY10 pKa = 9.13LKK12 pKa = 10.5SIMLPEE18 pKa = 4.74TGPASIPDD26 pKa = 5.16DD27 pKa = 3.12ITEE30 pKa = 3.99RR31 pKa = 11.84HH32 pKa = 5.89ILKK35 pKa = 10.07QEE37 pKa = 3.84TSSYY41 pKa = 9.6NLEE44 pKa = 4.05VSEE47 pKa = 4.58SGSGVLVCFPGAPGSRR63 pKa = 11.84IGAHH67 pKa = 5.45YY68 pKa = 9.97RR69 pKa = 11.84WNANQTGLEE78 pKa = 3.87FDD80 pKa = 3.19QWLEE84 pKa = 3.86TSQDD88 pKa = 3.07LKK90 pKa = 11.21KK91 pKa = 10.75AFNYY95 pKa = 9.81GRR97 pKa = 11.84LISRR101 pKa = 11.84KK102 pKa = 9.31YY103 pKa = 10.36DD104 pKa = 3.38IQSSTLPAGLYY115 pKa = 10.34ALNGTLNAATFEE127 pKa = 4.46GSLSEE132 pKa = 4.62VEE134 pKa = 4.23SLTYY138 pKa = 10.53NSLMSLTTNPQDD150 pKa = 3.31KK151 pKa = 10.9VNNQLVTKK159 pKa = 10.21GVTVLNLPTGFDD171 pKa = 3.17KK172 pKa = 10.97PYY174 pKa = 10.8VRR176 pKa = 11.84LEE178 pKa = 4.22DD179 pKa = 3.65EE180 pKa = 4.74TPQGLQSMNGAKK192 pKa = 8.9MRR194 pKa = 11.84CTAAIAPRR202 pKa = 11.84RR203 pKa = 11.84YY204 pKa = 10.24EE205 pKa = 3.63IDD207 pKa = 3.84LPSQRR212 pKa = 11.84LPPVPATGTLTTLYY226 pKa = 9.76EE227 pKa = 4.36GNADD231 pKa = 3.14IVNSTTVTGDD241 pKa = 3.01INFSLAEE248 pKa = 3.99QPADD252 pKa = 3.32EE253 pKa = 4.7TKK255 pKa = 10.68FDD257 pKa = 3.87FQLDD261 pKa = 3.72FMGLDD266 pKa = 3.12NDD268 pKa = 4.11VPVVTVVSSVLATNDD283 pKa = 2.83NYY285 pKa = 11.11RR286 pKa = 11.84GVSAKK291 pKa = 7.47MTQSIPTEE299 pKa = 4.45NITKK303 pKa = 10.03PITRR307 pKa = 11.84VKK309 pKa = 10.53LSYY312 pKa = 10.45KK313 pKa = 10.06INQQTAIGNVATLGTMGPASVSFSSGNGNVPGVLRR348 pKa = 11.84PITLVAYY355 pKa = 9.88EE356 pKa = 4.56KK357 pKa = 7.77MTPLSILTVAGVSNYY372 pKa = 9.4EE373 pKa = 4.44LIPNPEE379 pKa = 4.27LLKK382 pKa = 11.42NMVTRR387 pKa = 11.84YY388 pKa = 9.74GKK390 pKa = 10.45YY391 pKa = 10.22DD392 pKa = 3.71PEE394 pKa = 4.29GLNYY398 pKa = 10.95AKK400 pKa = 10.06MILSHH405 pKa = 7.46RR406 pKa = 11.84EE407 pKa = 3.56EE408 pKa = 4.78LDD410 pKa = 2.89IRR412 pKa = 11.84TVWRR416 pKa = 11.84TEE418 pKa = 3.79EE419 pKa = 3.98YY420 pKa = 10.51KK421 pKa = 10.88EE422 pKa = 3.75RR423 pKa = 11.84TRR425 pKa = 11.84VFNEE429 pKa = 3.48ITDD432 pKa = 4.37FSSDD436 pKa = 3.78LPTSKK441 pKa = 10.5AWGWRR446 pKa = 11.84DD447 pKa = 2.97IVRR450 pKa = 11.84GIRR453 pKa = 11.84KK454 pKa = 8.54VAAPVLSTLFPMAAPLIGMADD475 pKa = 3.49QFIGDD480 pKa = 3.9LTKK483 pKa = 10.75TNAAGGRR490 pKa = 11.84YY491 pKa = 9.37HH492 pKa = 8.3SMAAGGRR499 pKa = 11.84YY500 pKa = 9.4KK501 pKa = 10.73DD502 pKa = 4.16VLEE505 pKa = 4.56SWASGGPDD513 pKa = 3.01GKK515 pKa = 10.61FSRR518 pKa = 11.84ALKK521 pKa = 10.49NRR523 pKa = 11.84LEE525 pKa = 4.14SANYY529 pKa = 10.01EE530 pKa = 4.32EE531 pKa = 5.78VEE533 pKa = 4.58LPPPSKK539 pKa = 10.69GVIVPVVHH547 pKa = 5.83TVKK550 pKa = 10.46SAPGEE555 pKa = 4.17AFGSLAIIIPGEE567 pKa = 4.1YY568 pKa = 9.88PEE570 pKa = 5.82LLDD573 pKa = 5.53ANQQVLSHH581 pKa = 6.79FANDD585 pKa = 3.56TGSVWGIGEE594 pKa = 5.12DD595 pKa = 4.07IPFEE599 pKa = 4.38GDD601 pKa = 3.29NMCYY605 pKa = 9.05TALPLKK611 pKa = 9.86EE612 pKa = 3.89IKK614 pKa = 10.42RR615 pKa = 11.84NGNIVVEE622 pKa = 4.86KK623 pKa = 10.34IFAGPIMGPSAQLGLSLLVNDD644 pKa = 5.1IEE646 pKa = 5.31DD647 pKa = 3.96GVPRR651 pKa = 11.84MVFTGEE657 pKa = 3.79IADD660 pKa = 4.34DD661 pKa = 4.22EE662 pKa = 4.48EE663 pKa = 4.84TIIPICGVDD672 pKa = 3.33IKK674 pKa = 11.13AIAAHH679 pKa = 5.51EE680 pKa = 4.08QGLPLIGNQPGVDD693 pKa = 3.08EE694 pKa = 4.43EE695 pKa = 4.5VRR697 pKa = 11.84NTSLAAHH704 pKa = 7.67LIQTGTLPVQRR715 pKa = 11.84AKK717 pKa = 11.05GSNKK721 pKa = 9.2RR722 pKa = 11.84IKK724 pKa = 10.75YY725 pKa = 9.78LGEE728 pKa = 3.98LMASNASGMDD738 pKa = 3.43EE739 pKa = 3.92EE740 pKa = 5.05LQRR743 pKa = 11.84LLNATMARR751 pKa = 11.84AKK753 pKa = 9.75EE754 pKa = 4.04VQDD757 pKa = 3.54AEE759 pKa = 4.25IYY761 pKa = 10.62KK762 pKa = 10.24LLKK765 pKa = 10.5LMAWTRR771 pKa = 11.84KK772 pKa = 9.59NDD774 pKa = 3.94LTDD777 pKa = 5.05HH778 pKa = 5.96MYY780 pKa = 10.29EE781 pKa = 3.96WSKK784 pKa = 11.34EE785 pKa = 4.01DD786 pKa = 3.59PDD788 pKa = 3.72ALKK791 pKa = 10.49FGKK794 pKa = 10.38LISTPPKK801 pKa = 10.27HH802 pKa = 6.15PEE804 pKa = 3.87KK805 pKa = 10.57PKK807 pKa = 11.16GPDD810 pKa = 2.87QHH812 pKa = 6.6HH813 pKa = 6.05AQEE816 pKa = 4.23ARR818 pKa = 11.84ATRR821 pKa = 11.84ISLDD825 pKa = 3.32AVRR828 pKa = 11.84AGADD832 pKa = 3.26FATPEE837 pKa = 3.85WVALNNYY844 pKa = 8.8RR845 pKa = 11.84GPSPGQFKK853 pKa = 11.0YY854 pKa = 11.19YY855 pKa = 10.24LITGRR860 pKa = 11.84EE861 pKa = 4.09PEE863 pKa = 4.93PGDD866 pKa = 3.71EE867 pKa = 4.18YY868 pKa = 11.25EE869 pKa = 5.85DD870 pKa = 4.6YY871 pKa = 10.8IKK873 pKa = 10.78QPIVKK878 pKa = 8.42PTDD881 pKa = 3.21MNKK884 pKa = 9.11IRR886 pKa = 11.84RR887 pKa = 11.84LANSVYY893 pKa = 10.06GLPHH897 pKa = 6.09QEE899 pKa = 3.77PAPEE903 pKa = 3.88EE904 pKa = 4.51FYY906 pKa = 11.33DD907 pKa = 3.79AVAAVFAQNGGRR919 pKa = 11.84GPDD922 pKa = 3.35QDD924 pKa = 3.31QMQDD928 pKa = 3.13LRR930 pKa = 11.84EE931 pKa = 4.26LARR934 pKa = 11.84QMKK937 pKa = 9.58RR938 pKa = 11.84RR939 pKa = 11.84PRR941 pKa = 11.84NADD944 pKa = 2.72APRR947 pKa = 11.84RR948 pKa = 11.84TRR950 pKa = 11.84APAEE954 pKa = 3.76PAPPGRR960 pKa = 11.84SRR962 pKa = 11.84FTPSGDD968 pKa = 3.33NAEE971 pKa = 4.26VV972 pKa = 3.07
MM1 pKa = 6.89NTNKK5 pKa = 8.97ATATYY10 pKa = 9.13LKK12 pKa = 10.5SIMLPEE18 pKa = 4.74TGPASIPDD26 pKa = 5.16DD27 pKa = 3.12ITEE30 pKa = 3.99RR31 pKa = 11.84HH32 pKa = 5.89ILKK35 pKa = 10.07QEE37 pKa = 3.84TSSYY41 pKa = 9.6NLEE44 pKa = 4.05VSEE47 pKa = 4.58SGSGVLVCFPGAPGSRR63 pKa = 11.84IGAHH67 pKa = 5.45YY68 pKa = 9.97RR69 pKa = 11.84WNANQTGLEE78 pKa = 3.87FDD80 pKa = 3.19QWLEE84 pKa = 3.86TSQDD88 pKa = 3.07LKK90 pKa = 11.21KK91 pKa = 10.75AFNYY95 pKa = 9.81GRR97 pKa = 11.84LISRR101 pKa = 11.84KK102 pKa = 9.31YY103 pKa = 10.36DD104 pKa = 3.38IQSSTLPAGLYY115 pKa = 10.34ALNGTLNAATFEE127 pKa = 4.46GSLSEE132 pKa = 4.62VEE134 pKa = 4.23SLTYY138 pKa = 10.53NSLMSLTTNPQDD150 pKa = 3.31KK151 pKa = 10.9VNNQLVTKK159 pKa = 10.21GVTVLNLPTGFDD171 pKa = 3.17KK172 pKa = 10.97PYY174 pKa = 10.8VRR176 pKa = 11.84LEE178 pKa = 4.22DD179 pKa = 3.65EE180 pKa = 4.74TPQGLQSMNGAKK192 pKa = 8.9MRR194 pKa = 11.84CTAAIAPRR202 pKa = 11.84RR203 pKa = 11.84YY204 pKa = 10.24EE205 pKa = 3.63IDD207 pKa = 3.84LPSQRR212 pKa = 11.84LPPVPATGTLTTLYY226 pKa = 9.76EE227 pKa = 4.36GNADD231 pKa = 3.14IVNSTTVTGDD241 pKa = 3.01INFSLAEE248 pKa = 3.99QPADD252 pKa = 3.32EE253 pKa = 4.7TKK255 pKa = 10.68FDD257 pKa = 3.87FQLDD261 pKa = 3.72FMGLDD266 pKa = 3.12NDD268 pKa = 4.11VPVVTVVSSVLATNDD283 pKa = 2.83NYY285 pKa = 11.11RR286 pKa = 11.84GVSAKK291 pKa = 7.47MTQSIPTEE299 pKa = 4.45NITKK303 pKa = 10.03PITRR307 pKa = 11.84VKK309 pKa = 10.53LSYY312 pKa = 10.45KK313 pKa = 10.06INQQTAIGNVATLGTMGPASVSFSSGNGNVPGVLRR348 pKa = 11.84PITLVAYY355 pKa = 9.88EE356 pKa = 4.56KK357 pKa = 7.77MTPLSILTVAGVSNYY372 pKa = 9.4EE373 pKa = 4.44LIPNPEE379 pKa = 4.27LLKK382 pKa = 11.42NMVTRR387 pKa = 11.84YY388 pKa = 9.74GKK390 pKa = 10.45YY391 pKa = 10.22DD392 pKa = 3.71PEE394 pKa = 4.29GLNYY398 pKa = 10.95AKK400 pKa = 10.06MILSHH405 pKa = 7.46RR406 pKa = 11.84EE407 pKa = 3.56EE408 pKa = 4.78LDD410 pKa = 2.89IRR412 pKa = 11.84TVWRR416 pKa = 11.84TEE418 pKa = 3.79EE419 pKa = 3.98YY420 pKa = 10.51KK421 pKa = 10.88EE422 pKa = 3.75RR423 pKa = 11.84TRR425 pKa = 11.84VFNEE429 pKa = 3.48ITDD432 pKa = 4.37FSSDD436 pKa = 3.78LPTSKK441 pKa = 10.5AWGWRR446 pKa = 11.84DD447 pKa = 2.97IVRR450 pKa = 11.84GIRR453 pKa = 11.84KK454 pKa = 8.54VAAPVLSTLFPMAAPLIGMADD475 pKa = 3.49QFIGDD480 pKa = 3.9LTKK483 pKa = 10.75TNAAGGRR490 pKa = 11.84YY491 pKa = 9.37HH492 pKa = 8.3SMAAGGRR499 pKa = 11.84YY500 pKa = 9.4KK501 pKa = 10.73DD502 pKa = 4.16VLEE505 pKa = 4.56SWASGGPDD513 pKa = 3.01GKK515 pKa = 10.61FSRR518 pKa = 11.84ALKK521 pKa = 10.49NRR523 pKa = 11.84LEE525 pKa = 4.14SANYY529 pKa = 10.01EE530 pKa = 4.32EE531 pKa = 5.78VEE533 pKa = 4.58LPPPSKK539 pKa = 10.69GVIVPVVHH547 pKa = 5.83TVKK550 pKa = 10.46SAPGEE555 pKa = 4.17AFGSLAIIIPGEE567 pKa = 4.1YY568 pKa = 9.88PEE570 pKa = 5.82LLDD573 pKa = 5.53ANQQVLSHH581 pKa = 6.79FANDD585 pKa = 3.56TGSVWGIGEE594 pKa = 5.12DD595 pKa = 4.07IPFEE599 pKa = 4.38GDD601 pKa = 3.29NMCYY605 pKa = 9.05TALPLKK611 pKa = 9.86EE612 pKa = 3.89IKK614 pKa = 10.42RR615 pKa = 11.84NGNIVVEE622 pKa = 4.86KK623 pKa = 10.34IFAGPIMGPSAQLGLSLLVNDD644 pKa = 5.1IEE646 pKa = 5.31DD647 pKa = 3.96GVPRR651 pKa = 11.84MVFTGEE657 pKa = 3.79IADD660 pKa = 4.34DD661 pKa = 4.22EE662 pKa = 4.48EE663 pKa = 4.84TIIPICGVDD672 pKa = 3.33IKK674 pKa = 11.13AIAAHH679 pKa = 5.51EE680 pKa = 4.08QGLPLIGNQPGVDD693 pKa = 3.08EE694 pKa = 4.43EE695 pKa = 4.5VRR697 pKa = 11.84NTSLAAHH704 pKa = 7.67LIQTGTLPVQRR715 pKa = 11.84AKK717 pKa = 11.05GSNKK721 pKa = 9.2RR722 pKa = 11.84IKK724 pKa = 10.75YY725 pKa = 9.78LGEE728 pKa = 3.98LMASNASGMDD738 pKa = 3.43EE739 pKa = 3.92EE740 pKa = 5.05LQRR743 pKa = 11.84LLNATMARR751 pKa = 11.84AKK753 pKa = 9.75EE754 pKa = 4.04VQDD757 pKa = 3.54AEE759 pKa = 4.25IYY761 pKa = 10.62KK762 pKa = 10.24LLKK765 pKa = 10.5LMAWTRR771 pKa = 11.84KK772 pKa = 9.59NDD774 pKa = 3.94LTDD777 pKa = 5.05HH778 pKa = 5.96MYY780 pKa = 10.29EE781 pKa = 3.96WSKK784 pKa = 11.34EE785 pKa = 4.01DD786 pKa = 3.59PDD788 pKa = 3.72ALKK791 pKa = 10.49FGKK794 pKa = 10.38LISTPPKK801 pKa = 10.27HH802 pKa = 6.15PEE804 pKa = 3.87KK805 pKa = 10.57PKK807 pKa = 11.16GPDD810 pKa = 2.87QHH812 pKa = 6.6HH813 pKa = 6.05AQEE816 pKa = 4.23ARR818 pKa = 11.84ATRR821 pKa = 11.84ISLDD825 pKa = 3.32AVRR828 pKa = 11.84AGADD832 pKa = 3.26FATPEE837 pKa = 3.85WVALNNYY844 pKa = 8.8RR845 pKa = 11.84GPSPGQFKK853 pKa = 11.0YY854 pKa = 11.19YY855 pKa = 10.24LITGRR860 pKa = 11.84EE861 pKa = 4.09PEE863 pKa = 4.93PGDD866 pKa = 3.71EE867 pKa = 4.18YY868 pKa = 11.25EE869 pKa = 5.85DD870 pKa = 4.6YY871 pKa = 10.8IKK873 pKa = 10.78QPIVKK878 pKa = 8.42PTDD881 pKa = 3.21MNKK884 pKa = 9.11IRR886 pKa = 11.84RR887 pKa = 11.84LANSVYY893 pKa = 10.06GLPHH897 pKa = 6.09QEE899 pKa = 3.77PAPEE903 pKa = 3.88EE904 pKa = 4.51FYY906 pKa = 11.33DD907 pKa = 3.79AVAAVFAQNGGRR919 pKa = 11.84GPDD922 pKa = 3.35QDD924 pKa = 3.31QMQDD928 pKa = 3.13LRR930 pKa = 11.84EE931 pKa = 4.26LARR934 pKa = 11.84QMKK937 pKa = 9.58RR938 pKa = 11.84RR939 pKa = 11.84PRR941 pKa = 11.84NADD944 pKa = 2.72APRR947 pKa = 11.84RR948 pKa = 11.84TRR950 pKa = 11.84APAEE954 pKa = 3.76PAPPGRR960 pKa = 11.84SRR962 pKa = 11.84FTPSGDD968 pKa = 3.33NAEE971 pKa = 4.26VV972 pKa = 3.07
Molecular weight: 106.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q703G9|POLS_IPNVS Structural polyprotein OS=Infectious pancreatic necrosis virus (strain Sp) OX=11005 PE=1 SV=1
MM1 pKa = 7.67SDD3 pKa = 4.08IFNSPQNKK11 pKa = 9.77ASILNALMKK20 pKa = 9.7STQGDD25 pKa = 3.83VEE27 pKa = 4.64DD28 pKa = 3.71VLIPKK33 pKa = 10.06RR34 pKa = 11.84FRR36 pKa = 11.84PAKK39 pKa = 10.35DD40 pKa = 3.61PLDD43 pKa = 4.22SPQAAAAFLKK53 pKa = 8.53EE54 pKa = 3.81HH55 pKa = 7.62KK56 pKa = 10.37YY57 pKa = 10.63RR58 pKa = 11.84ILRR61 pKa = 11.84PRR63 pKa = 11.84AIPTMVEE70 pKa = 3.76IEE72 pKa = 3.98TDD74 pKa = 3.16AALPRR79 pKa = 11.84LAAMVDD85 pKa = 3.51DD86 pKa = 5.06GKK88 pKa = 11.05LKK90 pKa = 10.86EE91 pKa = 4.23MVNVPEE97 pKa = 4.55GTTAFYY103 pKa = 10.05PKK105 pKa = 9.82YY106 pKa = 10.15YY107 pKa = 9.64PFHH110 pKa = 7.17RR111 pKa = 11.84PDD113 pKa = 4.02HH114 pKa = 7.24DD115 pKa = 4.14DD116 pKa = 3.18VGTFGAPDD124 pKa = 3.15ITLLKK129 pKa = 10.52QLTFFLLEE137 pKa = 3.99NDD139 pKa = 4.02FPTGPEE145 pKa = 3.55TLRR148 pKa = 11.84QVRR151 pKa = 11.84EE152 pKa = 4.48AIATLQYY159 pKa = 11.35GSGSYY164 pKa = 10.01SGQLNRR170 pKa = 11.84LLAMKK175 pKa = 10.44GVATGRR181 pKa = 11.84NPNKK185 pKa = 9.57TPLAVGYY192 pKa = 8.5TNEE195 pKa = 3.64QMARR199 pKa = 11.84LMEE202 pKa = 4.05QTLPINPPKK211 pKa = 10.96NEE213 pKa = 4.26DD214 pKa = 3.58PDD216 pKa = 4.95LRR218 pKa = 11.84WAPSWLIQYY227 pKa = 8.7TGDD230 pKa = 3.31ASTDD234 pKa = 3.1KK235 pKa = 11.06SYY237 pKa = 11.24LPHH240 pKa = 5.43VTAKK244 pKa = 10.48SSAGLPYY251 pKa = 9.88IGKK254 pKa = 8.15TKK256 pKa = 10.95GDD258 pKa = 3.64TTAEE262 pKa = 4.03ALVLADD268 pKa = 4.42SFIRR272 pKa = 11.84DD273 pKa = 3.54LGKK276 pKa = 10.31AATSADD282 pKa = 3.33PGAAAKK288 pKa = 10.14KK289 pKa = 9.51VLSDD293 pKa = 3.77FWYY296 pKa = 10.37LSCGLLFPKK305 pKa = 10.51GEE307 pKa = 4.59RR308 pKa = 11.84YY309 pKa = 7.04TQKK312 pKa = 11.01DD313 pKa = 2.99WDD315 pKa = 4.02LKK317 pKa = 9.48TRR319 pKa = 11.84NIWSAPYY326 pKa = 7.69PTHH329 pKa = 7.06LLLSMVSSPVMDD341 pKa = 3.73EE342 pKa = 4.1SKK344 pKa = 11.34LNITNTQTPSLYY356 pKa = 10.6GFSPFHH362 pKa = 6.53GGINRR367 pKa = 11.84IMTIIRR373 pKa = 11.84EE374 pKa = 4.27HH375 pKa = 7.06LDD377 pKa = 3.36QEE379 pKa = 4.18QDD381 pKa = 3.18LVMIYY386 pKa = 10.95ADD388 pKa = 4.6NIYY391 pKa = 10.23ILQDD395 pKa = 3.45NTWYY399 pKa = 10.96SIDD402 pKa = 4.01LEE404 pKa = 4.4KK405 pKa = 11.54GEE407 pKa = 4.74ANCTPQHH414 pKa = 5.47MQAMMYY420 pKa = 10.23YY421 pKa = 9.91RR422 pKa = 11.84LTRR425 pKa = 11.84EE426 pKa = 3.81WTNEE430 pKa = 3.98DD431 pKa = 2.94GSPRR435 pKa = 11.84YY436 pKa = 9.89NPTWATFAMYY446 pKa = 9.92VGPSMVVDD454 pKa = 4.21STCLLMNLQLKK465 pKa = 6.82TTGQGSGNAFTFLNNHH481 pKa = 6.13LMSTIVVAEE490 pKa = 3.65WHH492 pKa = 6.25KK493 pKa = 11.05AGRR496 pKa = 11.84PNPMSKK502 pKa = 10.45EE503 pKa = 3.95FMDD506 pKa = 5.28LEE508 pKa = 4.64AKK510 pKa = 9.88TGINFKK516 pKa = 10.2IEE518 pKa = 4.2RR519 pKa = 11.84EE520 pKa = 4.18LKK522 pKa = 9.94DD523 pKa = 3.35LRR525 pKa = 11.84SIIMEE530 pKa = 4.43AVDD533 pKa = 3.58TAPLDD538 pKa = 4.01GYY540 pKa = 11.04LADD543 pKa = 5.56GSDD546 pKa = 3.95LPPRR550 pKa = 11.84VPGKK554 pKa = 10.37AVEE557 pKa = 4.12LDD559 pKa = 3.63LLGWSAVYY567 pKa = 10.23SRR569 pKa = 11.84QLEE572 pKa = 4.22MFVPVLEE579 pKa = 4.05NEE581 pKa = 4.02RR582 pKa = 11.84LIASVAYY589 pKa = 9.32PKK591 pKa = 10.89GLEE594 pKa = 4.09NKK596 pKa = 10.05SLARR600 pKa = 11.84KK601 pKa = 8.88PGAEE605 pKa = 3.3IAYY608 pKa = 9.8QIVRR612 pKa = 11.84YY613 pKa = 8.57EE614 pKa = 4.38AIRR617 pKa = 11.84LIGGWNNPLIEE628 pKa = 4.13TAAKK632 pKa = 10.34HH633 pKa = 5.12MSLDD637 pKa = 3.04KK638 pKa = 10.75RR639 pKa = 11.84KK640 pKa = 8.93RR641 pKa = 11.84LEE643 pKa = 3.9VKK645 pKa = 10.72GIDD648 pKa = 3.35VTGFLDD654 pKa = 3.56DD655 pKa = 3.97WNTMSEE661 pKa = 4.18FGGDD665 pKa = 3.55LEE667 pKa = 5.43GISLTAPLTNQTLLDD682 pKa = 3.63INTPEE687 pKa = 4.05TEE689 pKa = 4.02FDD691 pKa = 3.79VKK693 pKa = 10.77DD694 pKa = 3.84RR695 pKa = 11.84PPTPRR700 pKa = 11.84SPGKK704 pKa = 8.25TLAEE708 pKa = 3.92VTAAITSGTYY718 pKa = 9.95KK719 pKa = 10.54DD720 pKa = 3.78PKK722 pKa = 8.57SAVWRR727 pKa = 11.84LLDD730 pKa = 3.27QRR732 pKa = 11.84TKK734 pKa = 11.18LRR736 pKa = 11.84VSTLRR741 pKa = 11.84DD742 pKa = 3.04HH743 pKa = 7.15AHH745 pKa = 6.38ALKK748 pKa = 9.97PAASTSDD755 pKa = 3.35FWGDD759 pKa = 3.15ATEE762 pKa = 4.83EE763 pKa = 4.01LAEE766 pKa = 3.99QQQLLMKK773 pKa = 10.59ANNLLKK779 pKa = 10.81SSLTEE784 pKa = 3.57ARR786 pKa = 11.84EE787 pKa = 3.98ALEE790 pKa = 4.35TVQSDD795 pKa = 3.89KK796 pKa = 10.91IISGKK801 pKa = 8.33TSPEE805 pKa = 4.1KK806 pKa = 11.11NPGTAANPVVAYY818 pKa = 10.43GEE820 pKa = 4.16FSEE823 pKa = 5.61KK824 pKa = 10.33IPLTPTQKK832 pKa = 10.69KK833 pKa = 7.32NAKK836 pKa = 9.43RR837 pKa = 11.84RR838 pKa = 11.84EE839 pKa = 3.99KK840 pKa = 10.41QRR842 pKa = 11.84RR843 pKa = 11.84NN844 pKa = 3.14
MM1 pKa = 7.67SDD3 pKa = 4.08IFNSPQNKK11 pKa = 9.77ASILNALMKK20 pKa = 9.7STQGDD25 pKa = 3.83VEE27 pKa = 4.64DD28 pKa = 3.71VLIPKK33 pKa = 10.06RR34 pKa = 11.84FRR36 pKa = 11.84PAKK39 pKa = 10.35DD40 pKa = 3.61PLDD43 pKa = 4.22SPQAAAAFLKK53 pKa = 8.53EE54 pKa = 3.81HH55 pKa = 7.62KK56 pKa = 10.37YY57 pKa = 10.63RR58 pKa = 11.84ILRR61 pKa = 11.84PRR63 pKa = 11.84AIPTMVEE70 pKa = 3.76IEE72 pKa = 3.98TDD74 pKa = 3.16AALPRR79 pKa = 11.84LAAMVDD85 pKa = 3.51DD86 pKa = 5.06GKK88 pKa = 11.05LKK90 pKa = 10.86EE91 pKa = 4.23MVNVPEE97 pKa = 4.55GTTAFYY103 pKa = 10.05PKK105 pKa = 9.82YY106 pKa = 10.15YY107 pKa = 9.64PFHH110 pKa = 7.17RR111 pKa = 11.84PDD113 pKa = 4.02HH114 pKa = 7.24DD115 pKa = 4.14DD116 pKa = 3.18VGTFGAPDD124 pKa = 3.15ITLLKK129 pKa = 10.52QLTFFLLEE137 pKa = 3.99NDD139 pKa = 4.02FPTGPEE145 pKa = 3.55TLRR148 pKa = 11.84QVRR151 pKa = 11.84EE152 pKa = 4.48AIATLQYY159 pKa = 11.35GSGSYY164 pKa = 10.01SGQLNRR170 pKa = 11.84LLAMKK175 pKa = 10.44GVATGRR181 pKa = 11.84NPNKK185 pKa = 9.57TPLAVGYY192 pKa = 8.5TNEE195 pKa = 3.64QMARR199 pKa = 11.84LMEE202 pKa = 4.05QTLPINPPKK211 pKa = 10.96NEE213 pKa = 4.26DD214 pKa = 3.58PDD216 pKa = 4.95LRR218 pKa = 11.84WAPSWLIQYY227 pKa = 8.7TGDD230 pKa = 3.31ASTDD234 pKa = 3.1KK235 pKa = 11.06SYY237 pKa = 11.24LPHH240 pKa = 5.43VTAKK244 pKa = 10.48SSAGLPYY251 pKa = 9.88IGKK254 pKa = 8.15TKK256 pKa = 10.95GDD258 pKa = 3.64TTAEE262 pKa = 4.03ALVLADD268 pKa = 4.42SFIRR272 pKa = 11.84DD273 pKa = 3.54LGKK276 pKa = 10.31AATSADD282 pKa = 3.33PGAAAKK288 pKa = 10.14KK289 pKa = 9.51VLSDD293 pKa = 3.77FWYY296 pKa = 10.37LSCGLLFPKK305 pKa = 10.51GEE307 pKa = 4.59RR308 pKa = 11.84YY309 pKa = 7.04TQKK312 pKa = 11.01DD313 pKa = 2.99WDD315 pKa = 4.02LKK317 pKa = 9.48TRR319 pKa = 11.84NIWSAPYY326 pKa = 7.69PTHH329 pKa = 7.06LLLSMVSSPVMDD341 pKa = 3.73EE342 pKa = 4.1SKK344 pKa = 11.34LNITNTQTPSLYY356 pKa = 10.6GFSPFHH362 pKa = 6.53GGINRR367 pKa = 11.84IMTIIRR373 pKa = 11.84EE374 pKa = 4.27HH375 pKa = 7.06LDD377 pKa = 3.36QEE379 pKa = 4.18QDD381 pKa = 3.18LVMIYY386 pKa = 10.95ADD388 pKa = 4.6NIYY391 pKa = 10.23ILQDD395 pKa = 3.45NTWYY399 pKa = 10.96SIDD402 pKa = 4.01LEE404 pKa = 4.4KK405 pKa = 11.54GEE407 pKa = 4.74ANCTPQHH414 pKa = 5.47MQAMMYY420 pKa = 10.23YY421 pKa = 9.91RR422 pKa = 11.84LTRR425 pKa = 11.84EE426 pKa = 3.81WTNEE430 pKa = 3.98DD431 pKa = 2.94GSPRR435 pKa = 11.84YY436 pKa = 9.89NPTWATFAMYY446 pKa = 9.92VGPSMVVDD454 pKa = 4.21STCLLMNLQLKK465 pKa = 6.82TTGQGSGNAFTFLNNHH481 pKa = 6.13LMSTIVVAEE490 pKa = 3.65WHH492 pKa = 6.25KK493 pKa = 11.05AGRR496 pKa = 11.84PNPMSKK502 pKa = 10.45EE503 pKa = 3.95FMDD506 pKa = 5.28LEE508 pKa = 4.64AKK510 pKa = 9.88TGINFKK516 pKa = 10.2IEE518 pKa = 4.2RR519 pKa = 11.84EE520 pKa = 4.18LKK522 pKa = 9.94DD523 pKa = 3.35LRR525 pKa = 11.84SIIMEE530 pKa = 4.43AVDD533 pKa = 3.58TAPLDD538 pKa = 4.01GYY540 pKa = 11.04LADD543 pKa = 5.56GSDD546 pKa = 3.95LPPRR550 pKa = 11.84VPGKK554 pKa = 10.37AVEE557 pKa = 4.12LDD559 pKa = 3.63LLGWSAVYY567 pKa = 10.23SRR569 pKa = 11.84QLEE572 pKa = 4.22MFVPVLEE579 pKa = 4.05NEE581 pKa = 4.02RR582 pKa = 11.84LIASVAYY589 pKa = 9.32PKK591 pKa = 10.89GLEE594 pKa = 4.09NKK596 pKa = 10.05SLARR600 pKa = 11.84KK601 pKa = 8.88PGAEE605 pKa = 3.3IAYY608 pKa = 9.8QIVRR612 pKa = 11.84YY613 pKa = 8.57EE614 pKa = 4.38AIRR617 pKa = 11.84LIGGWNNPLIEE628 pKa = 4.13TAAKK632 pKa = 10.34HH633 pKa = 5.12MSLDD637 pKa = 3.04KK638 pKa = 10.75RR639 pKa = 11.84KK640 pKa = 8.93RR641 pKa = 11.84LEE643 pKa = 3.9VKK645 pKa = 10.72GIDD648 pKa = 3.35VTGFLDD654 pKa = 3.56DD655 pKa = 3.97WNTMSEE661 pKa = 4.18FGGDD665 pKa = 3.55LEE667 pKa = 5.43GISLTAPLTNQTLLDD682 pKa = 3.63INTPEE687 pKa = 4.05TEE689 pKa = 4.02FDD691 pKa = 3.79VKK693 pKa = 10.77DD694 pKa = 3.84RR695 pKa = 11.84PPTPRR700 pKa = 11.84SPGKK704 pKa = 8.25TLAEE708 pKa = 3.92VTAAITSGTYY718 pKa = 9.95KK719 pKa = 10.54DD720 pKa = 3.78PKK722 pKa = 8.57SAVWRR727 pKa = 11.84LLDD730 pKa = 3.27QRR732 pKa = 11.84TKK734 pKa = 11.18LRR736 pKa = 11.84VSTLRR741 pKa = 11.84DD742 pKa = 3.04HH743 pKa = 7.15AHH745 pKa = 6.38ALKK748 pKa = 9.97PAASTSDD755 pKa = 3.35FWGDD759 pKa = 3.15ATEE762 pKa = 4.83EE763 pKa = 4.01LAEE766 pKa = 3.99QQQLLMKK773 pKa = 10.59ANNLLKK779 pKa = 10.81SSLTEE784 pKa = 3.57ARR786 pKa = 11.84EE787 pKa = 3.98ALEE790 pKa = 4.35TVQSDD795 pKa = 3.89KK796 pKa = 10.91IISGKK801 pKa = 8.33TSPEE805 pKa = 4.1KK806 pKa = 11.11NPGTAANPVVAYY818 pKa = 10.43GEE820 pKa = 4.16FSEE823 pKa = 5.61KK824 pKa = 10.33IPLTPTQKK832 pKa = 10.69KK833 pKa = 7.32NAKK836 pKa = 9.43RR837 pKa = 11.84RR838 pKa = 11.84EE839 pKa = 3.99KK840 pKa = 10.41QRR842 pKa = 11.84RR843 pKa = 11.84NN844 pKa = 3.14
Molecular weight: 94.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2660 |
844 |
972 |
886.7 |
98.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.571 ± 0.119 | 0.376 ± 0.015 |
6.053 ± 0.22 | 6.316 ± 0.204 |
2.857 ± 0.078 | 6.617 ± 0.47 |
1.466 ± 0.055 | 5.0 ± 0.195 |
6.053 ± 0.396 | 9.812 ± 0.509 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.895 ± 0.139 | 4.85 ± 0.221 |
7.03 ± 0.118 | 3.534 ± 0.037 |
5.113 ± 0.123 | 6.203 ± 0.032 |
7.368 ± 0.249 | 5.15 ± 0.392 |
1.429 ± 0.172 | 3.308 ± 0.05 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
