
Sewage-associated gemycircularvirus 9
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus sewopo5; Sewage derived gemycircularvirus 5
Average proteome isoelectric point is 7.44
Get precalculated fractions of proteins
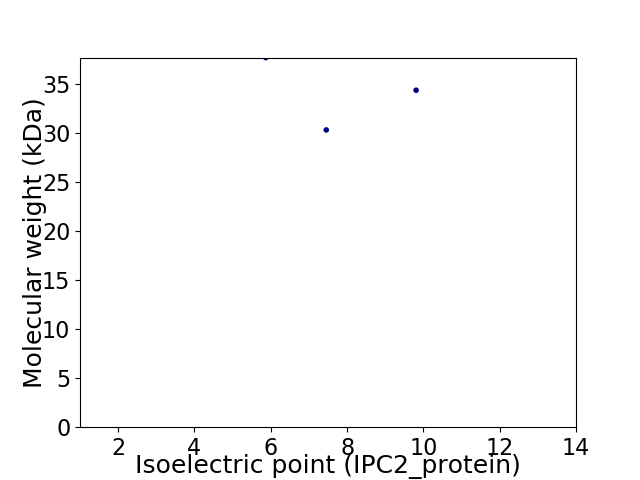
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0A7CL54|A0A0A7CL54_9VIRU Capsid protein OS=Sewage-associated gemycircularvirus 9 OX=1519404 PE=4 SV=1
MM1 pKa = 7.91PFNLHH6 pKa = 6.06CRR8 pKa = 11.84YY9 pKa = 10.45ALLTYY14 pKa = 8.89AQCGDD19 pKa = 4.5LSPTAVGEE27 pKa = 4.16FFDD30 pKa = 3.66NLGYY34 pKa = 10.65KK35 pKa = 9.9SVIGRR40 pKa = 11.84EE41 pKa = 3.77NHH43 pKa = 6.65ADD45 pKa = 3.51GGVHH49 pKa = 5.94LHH51 pKa = 6.49CFVDD55 pKa = 4.74FGRR58 pKa = 11.84KK59 pKa = 7.94RR60 pKa = 11.84RR61 pKa = 11.84FRR63 pKa = 11.84RR64 pKa = 11.84ARR66 pKa = 11.84VFDD69 pKa = 3.33IEE71 pKa = 4.14NRR73 pKa = 11.84HH74 pKa = 6.13PNVEE78 pKa = 4.11PSRR81 pKa = 11.84GTPEE85 pKa = 4.4KK86 pKa = 10.5GWDD89 pKa = 3.54YY90 pKa = 11.42AVKK93 pKa = 10.71DD94 pKa = 3.87GDD96 pKa = 3.36ICYY99 pKa = 10.27QSLDD103 pKa = 3.56RR104 pKa = 11.84PRR106 pKa = 11.84EE107 pKa = 3.95SSPSGGSNGGTRR119 pKa = 11.84DD120 pKa = 2.86KK121 pKa = 10.3WASITGASDD130 pKa = 5.15RR131 pKa = 11.84EE132 pKa = 4.38AFWDD136 pKa = 4.24LVHH139 pKa = 7.02EE140 pKa = 5.15LDD142 pKa = 4.48PKK144 pKa = 10.83SAACCFTQLQKK155 pKa = 11.05YY156 pKa = 8.98CDD158 pKa = 3.24WKK160 pKa = 10.42FAPRR164 pKa = 11.84TPEE167 pKa = 3.8YY168 pKa = 9.78EE169 pKa = 4.24SPGGLEE175 pKa = 4.46FIGGGLDD182 pKa = 5.38GRR184 pKa = 11.84DD185 pKa = 3.26DD186 pKa = 3.68WLLQSGIGGRR196 pKa = 11.84EE197 pKa = 3.73AFVGEE202 pKa = 4.26LVCRR206 pKa = 11.84RR207 pKa = 11.84CMSICVYY214 pKa = 10.83GEE216 pKa = 3.81SRR218 pKa = 11.84TGKK221 pKa = 7.35TLWARR226 pKa = 11.84SLGPHH231 pKa = 7.14IYY233 pKa = 10.33CVGLVSGDD241 pKa = 3.51EE242 pKa = 4.16CMKK245 pKa = 11.05ASTADD250 pKa = 3.43YY251 pKa = 11.18AVFDD255 pKa = 5.43DD256 pKa = 3.7IRR258 pKa = 11.84GGIKK262 pKa = 9.99FFPSFKK268 pKa = 10.33EE269 pKa = 3.83WLGCQAWVSVKK280 pKa = 10.37CLYY283 pKa = 10.51RR284 pKa = 11.84EE285 pKa = 4.02PKK287 pKa = 8.81LVKK290 pKa = 9.08WGKK293 pKa = 9.13PSIWLSNTDD302 pKa = 3.02PRR304 pKa = 11.84DD305 pKa = 3.42YY306 pKa = 10.79MEE308 pKa = 5.32NSDD311 pKa = 4.57IDD313 pKa = 3.87WMNKK317 pKa = 7.88NCIFVEE323 pKa = 4.34VNSPIFRR330 pKa = 11.84ANTEE334 pKa = 3.84
MM1 pKa = 7.91PFNLHH6 pKa = 6.06CRR8 pKa = 11.84YY9 pKa = 10.45ALLTYY14 pKa = 8.89AQCGDD19 pKa = 4.5LSPTAVGEE27 pKa = 4.16FFDD30 pKa = 3.66NLGYY34 pKa = 10.65KK35 pKa = 9.9SVIGRR40 pKa = 11.84EE41 pKa = 3.77NHH43 pKa = 6.65ADD45 pKa = 3.51GGVHH49 pKa = 5.94LHH51 pKa = 6.49CFVDD55 pKa = 4.74FGRR58 pKa = 11.84KK59 pKa = 7.94RR60 pKa = 11.84RR61 pKa = 11.84FRR63 pKa = 11.84RR64 pKa = 11.84ARR66 pKa = 11.84VFDD69 pKa = 3.33IEE71 pKa = 4.14NRR73 pKa = 11.84HH74 pKa = 6.13PNVEE78 pKa = 4.11PSRR81 pKa = 11.84GTPEE85 pKa = 4.4KK86 pKa = 10.5GWDD89 pKa = 3.54YY90 pKa = 11.42AVKK93 pKa = 10.71DD94 pKa = 3.87GDD96 pKa = 3.36ICYY99 pKa = 10.27QSLDD103 pKa = 3.56RR104 pKa = 11.84PRR106 pKa = 11.84EE107 pKa = 3.95SSPSGGSNGGTRR119 pKa = 11.84DD120 pKa = 2.86KK121 pKa = 10.3WASITGASDD130 pKa = 5.15RR131 pKa = 11.84EE132 pKa = 4.38AFWDD136 pKa = 4.24LVHH139 pKa = 7.02EE140 pKa = 5.15LDD142 pKa = 4.48PKK144 pKa = 10.83SAACCFTQLQKK155 pKa = 11.05YY156 pKa = 8.98CDD158 pKa = 3.24WKK160 pKa = 10.42FAPRR164 pKa = 11.84TPEE167 pKa = 3.8YY168 pKa = 9.78EE169 pKa = 4.24SPGGLEE175 pKa = 4.46FIGGGLDD182 pKa = 5.38GRR184 pKa = 11.84DD185 pKa = 3.26DD186 pKa = 3.68WLLQSGIGGRR196 pKa = 11.84EE197 pKa = 3.73AFVGEE202 pKa = 4.26LVCRR206 pKa = 11.84RR207 pKa = 11.84CMSICVYY214 pKa = 10.83GEE216 pKa = 3.81SRR218 pKa = 11.84TGKK221 pKa = 7.35TLWARR226 pKa = 11.84SLGPHH231 pKa = 7.14IYY233 pKa = 10.33CVGLVSGDD241 pKa = 3.51EE242 pKa = 4.16CMKK245 pKa = 11.05ASTADD250 pKa = 3.43YY251 pKa = 11.18AVFDD255 pKa = 5.43DD256 pKa = 3.7IRR258 pKa = 11.84GGIKK262 pKa = 9.99FFPSFKK268 pKa = 10.33EE269 pKa = 3.83WLGCQAWVSVKK280 pKa = 10.37CLYY283 pKa = 10.51RR284 pKa = 11.84EE285 pKa = 4.02PKK287 pKa = 8.81LVKK290 pKa = 9.08WGKK293 pKa = 9.13PSIWLSNTDD302 pKa = 3.02PRR304 pKa = 11.84DD305 pKa = 3.42YY306 pKa = 10.79MEE308 pKa = 5.32NSDD311 pKa = 4.57IDD313 pKa = 3.87WMNKK317 pKa = 7.88NCIFVEE323 pKa = 4.34VNSPIFRR330 pKa = 11.84ANTEE334 pKa = 3.84
Molecular weight: 37.72 kDa
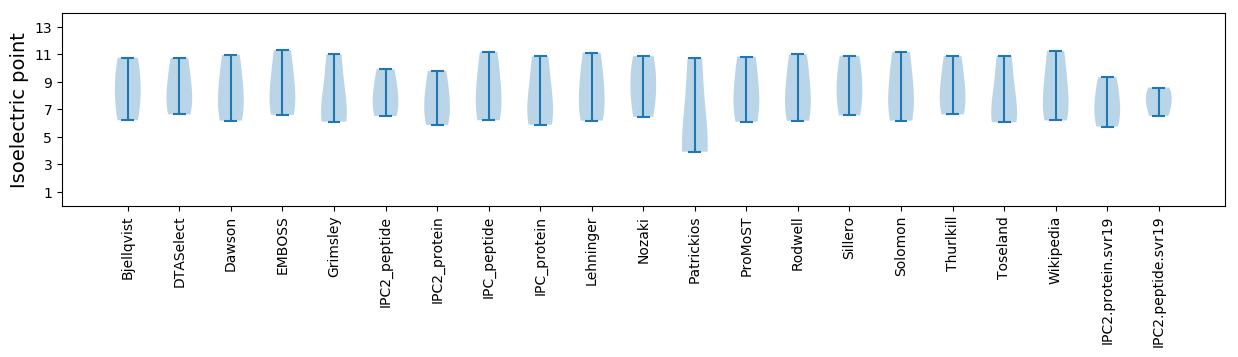
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7CL72|A0A0A7CL72_9VIRU RepA OS=Sewage-associated gemycircularvirus 9 OX=1519404 PE=3 SV=1
MM1 pKa = 7.28RR2 pKa = 11.84RR3 pKa = 11.84SRR5 pKa = 11.84RR6 pKa = 11.84FPAKK10 pKa = 9.97RR11 pKa = 11.84RR12 pKa = 11.84FTRR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 5.0TTYY20 pKa = 8.83RR21 pKa = 11.84RR22 pKa = 11.84PRR24 pKa = 11.84RR25 pKa = 11.84TITKK29 pKa = 8.6RR30 pKa = 11.84SYY32 pKa = 9.81RR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84PMRR39 pKa = 11.84RR40 pKa = 11.84MTRR43 pKa = 11.84KK44 pKa = 10.11SILNTTSTKK53 pKa = 10.46KK54 pKa = 10.04RR55 pKa = 11.84DD56 pKa = 3.25TMIAGNTFPALPEE69 pKa = 3.95NVGALTVTSDD79 pKa = 3.52VPAVLLWCPTARR91 pKa = 11.84QLADD95 pKa = 3.23TRR97 pKa = 11.84SALSTMPVNAQRR109 pKa = 11.84LSSTPFIVGLKK120 pKa = 7.67EE121 pKa = 4.11TITIRR126 pKa = 11.84TDD128 pKa = 2.9TSNAWRR134 pKa = 11.84WRR136 pKa = 11.84RR137 pKa = 11.84IVFFLKK143 pKa = 10.41GGPPGFTDD151 pKa = 2.87VGDD154 pKa = 3.69INRR157 pKa = 11.84VYY159 pKa = 11.13SPVNAGTDD167 pKa = 3.4FDD169 pKa = 4.46TYY171 pKa = 11.28QRR173 pKa = 11.84TATALPFALVADD185 pKa = 4.81VYY187 pKa = 11.65ASIFRR192 pKa = 11.84GFGINNVSATPQDD205 pKa = 3.27WLDD208 pKa = 4.92PITAPIDD215 pKa = 3.46TSRR218 pKa = 11.84ITPVYY223 pKa = 10.78DD224 pKa = 2.99HH225 pKa = 6.42TTSIRR230 pKa = 11.84SGNDD234 pKa = 2.58TGVVKK239 pKa = 8.26TTTSGTVSGRR249 pKa = 11.84HH250 pKa = 4.51STNNDD255 pKa = 3.07SEE257 pKa = 4.5IGGNMLSSAFSTHH270 pKa = 6.48FKK272 pKa = 10.08PGCGDD277 pKa = 3.68MYY279 pKa = 11.48VMDD282 pKa = 5.41IIIGNSAEE290 pKa = 4.32SSDD293 pKa = 3.87ALQFLPTSTLYY304 pKa = 9.65WHH306 pKa = 7.06EE307 pKa = 4.2KK308 pKa = 9.32
MM1 pKa = 7.28RR2 pKa = 11.84RR3 pKa = 11.84SRR5 pKa = 11.84RR6 pKa = 11.84FPAKK10 pKa = 9.97RR11 pKa = 11.84RR12 pKa = 11.84FTRR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 5.0TTYY20 pKa = 8.83RR21 pKa = 11.84RR22 pKa = 11.84PRR24 pKa = 11.84RR25 pKa = 11.84TITKK29 pKa = 8.6RR30 pKa = 11.84SYY32 pKa = 9.81RR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84PMRR39 pKa = 11.84RR40 pKa = 11.84MTRR43 pKa = 11.84KK44 pKa = 10.11SILNTTSTKK53 pKa = 10.46KK54 pKa = 10.04RR55 pKa = 11.84DD56 pKa = 3.25TMIAGNTFPALPEE69 pKa = 3.95NVGALTVTSDD79 pKa = 3.52VPAVLLWCPTARR91 pKa = 11.84QLADD95 pKa = 3.23TRR97 pKa = 11.84SALSTMPVNAQRR109 pKa = 11.84LSSTPFIVGLKK120 pKa = 7.67EE121 pKa = 4.11TITIRR126 pKa = 11.84TDD128 pKa = 2.9TSNAWRR134 pKa = 11.84WRR136 pKa = 11.84RR137 pKa = 11.84IVFFLKK143 pKa = 10.41GGPPGFTDD151 pKa = 2.87VGDD154 pKa = 3.69INRR157 pKa = 11.84VYY159 pKa = 11.13SPVNAGTDD167 pKa = 3.4FDD169 pKa = 4.46TYY171 pKa = 11.28QRR173 pKa = 11.84TATALPFALVADD185 pKa = 4.81VYY187 pKa = 11.65ASIFRR192 pKa = 11.84GFGINNVSATPQDD205 pKa = 3.27WLDD208 pKa = 4.92PITAPIDD215 pKa = 3.46TSRR218 pKa = 11.84ITPVYY223 pKa = 10.78DD224 pKa = 2.99HH225 pKa = 6.42TTSIRR230 pKa = 11.84SGNDD234 pKa = 2.58TGVVKK239 pKa = 8.26TTTSGTVSGRR249 pKa = 11.84HH250 pKa = 4.51STNNDD255 pKa = 3.07SEE257 pKa = 4.5IGGNMLSSAFSTHH270 pKa = 6.48FKK272 pKa = 10.08PGCGDD277 pKa = 3.68MYY279 pKa = 11.48VMDD282 pKa = 5.41IIIGNSAEE290 pKa = 4.32SSDD293 pKa = 3.87ALQFLPTSTLYY304 pKa = 9.65WHH306 pKa = 7.06EE307 pKa = 4.2KK308 pKa = 9.32
Molecular weight: 34.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
914 |
272 |
334 |
304.7 |
34.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.127 ± 0.276 | 3.063 ± 0.954 |
6.893 ± 0.412 | 4.158 ± 1.043 |
4.814 ± 0.256 | 9.519 ± 1.39 |
1.969 ± 0.279 | 4.705 ± 0.444 |
4.158 ± 0.412 | 7.002 ± 0.819 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.86 ± 0.288 | 3.611 ± 0.426 |
5.799 ± 0.314 | 1.969 ± 0.221 |
9.3 ± 0.814 | 7.44 ± 0.699 |
6.783 ± 2.546 | 5.252 ± 0.439 |
2.298 ± 0.441 | 3.282 ± 0.267 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
