
Mariniblastus fucicola
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae; Mariniblastus
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
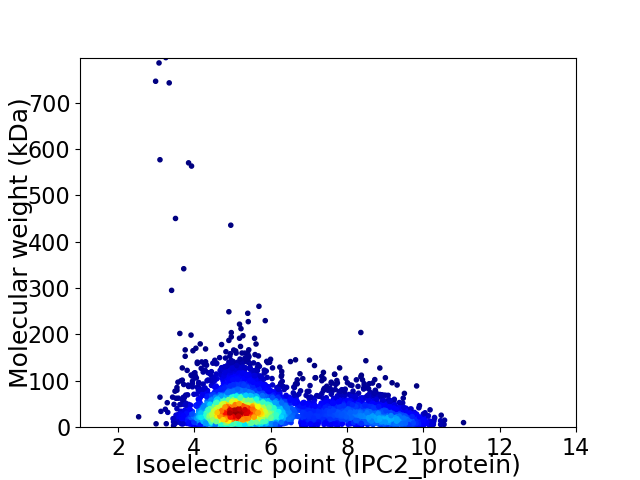
Virtual 2D-PAGE plot for 5083 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B9P529|A0A5B9P529_9BACT Trigger factor OS=Mariniblastus fucicola OX=980251 GN=tig PE=3 SV=1
MM1 pKa = 7.26NRR3 pKa = 11.84RR4 pKa = 11.84IKK6 pKa = 10.69SKK8 pKa = 10.89SRR10 pKa = 11.84FQSYY14 pKa = 7.71QQLEE18 pKa = 4.45AKK20 pKa = 10.12NLLAAIYY27 pKa = 8.94HH28 pKa = 6.0NPDD31 pKa = 2.87SGILYY36 pKa = 9.97IAGDD40 pKa = 3.34SGANVGQVSVSGDD53 pKa = 3.34NVVAQIDD60 pKa = 4.1GQSYY64 pKa = 10.56SGASDD69 pKa = 3.87DD70 pKa = 3.63VTDD73 pKa = 5.41IIFIGYY79 pKa = 9.56DD80 pKa = 3.39GNDD83 pKa = 3.19TFTNLSDD90 pKa = 5.01LDD92 pKa = 3.75SSMYY96 pKa = 10.72GHH98 pKa = 7.91LGNDD102 pKa = 3.43TLTGGGGDD110 pKa = 3.95DD111 pKa = 4.02NLVGGAGNDD120 pKa = 3.94TIQGLDD126 pKa = 3.31GDD128 pKa = 4.41DD129 pKa = 4.44RR130 pKa = 11.84IVGADD135 pKa = 3.59GNDD138 pKa = 3.5TLEE141 pKa = 4.81GGDD144 pKa = 3.66GEE146 pKa = 4.98DD147 pKa = 4.95RR148 pKa = 11.84IFGTAGTNTIYY159 pKa = 10.91GGAGDD164 pKa = 3.77DD165 pKa = 3.97TIYY168 pKa = 10.96GSEE171 pKa = 4.51DD172 pKa = 3.21ADD174 pKa = 4.55EE175 pKa = 5.1IYY177 pKa = 11.28GDD179 pKa = 3.66AGADD183 pKa = 3.34KK184 pKa = 10.33IYY186 pKa = 11.1ALGGDD191 pKa = 4.51DD192 pKa = 4.66ILYY195 pKa = 8.98TGTGGVEE202 pKa = 3.7GGSFDD207 pKa = 4.78EE208 pKa = 5.41GDD210 pKa = 3.26LAMGHH215 pKa = 6.34GGDD218 pKa = 4.12DD219 pKa = 3.41EE220 pKa = 6.02FYY222 pKa = 11.1GSTGLNIFYY231 pKa = 10.67GGDD234 pKa = 3.42GDD236 pKa = 5.55DD237 pKa = 3.74IMVGGSGEE245 pKa = 4.08NRR247 pKa = 11.84MHH249 pKa = 6.35GQAGDD254 pKa = 3.62DD255 pKa = 3.7TLTGGEE261 pKa = 4.49KK262 pKa = 10.17GDD264 pKa = 3.91YY265 pKa = 7.46MTGADD270 pKa = 4.9GDD272 pKa = 4.07DD273 pKa = 3.74TFDD276 pKa = 3.32GRR278 pKa = 11.84GGVDD282 pKa = 3.42YY283 pKa = 11.28YY284 pKa = 11.84NVGDD288 pKa = 3.99GSDD291 pKa = 3.73TIVYY295 pKa = 8.42SQSYY299 pKa = 9.08VPTDD303 pKa = 3.05VHH305 pKa = 5.17VTNNGGTAEE314 pKa = 3.98TRR316 pKa = 11.84IQNEE320 pKa = 4.3LVSNATWVQFADD332 pKa = 3.85RR333 pKa = 11.84TISADD338 pKa = 3.0QAYY341 pKa = 10.06YY342 pKa = 9.42LTVDD346 pKa = 3.42EE347 pKa = 5.31TNFSSLNGYY356 pKa = 8.95RR357 pKa = 11.84VSSSVGFVSKK367 pKa = 10.65PSDD370 pKa = 3.51LADD373 pKa = 4.02FAYY376 pKa = 10.45NWSLQMASAGTLSHH390 pKa = 5.8STEE393 pKa = 3.87SDD395 pKa = 3.14RR396 pKa = 11.84ASLYY400 pKa = 10.77VGDD403 pKa = 4.45RR404 pKa = 11.84SAVGEE409 pKa = 4.14NVAMVSDD416 pKa = 4.55TGQSDD421 pKa = 3.96VQIANYY427 pKa = 9.47FLSLWQSSSAHH438 pKa = 5.92NSNLLNSNYY447 pKa = 9.78QEE449 pKa = 3.96VGIGIVKK456 pKa = 10.46SGGAWWATQIFMGG469 pKa = 4.72
MM1 pKa = 7.26NRR3 pKa = 11.84RR4 pKa = 11.84IKK6 pKa = 10.69SKK8 pKa = 10.89SRR10 pKa = 11.84FQSYY14 pKa = 7.71QQLEE18 pKa = 4.45AKK20 pKa = 10.12NLLAAIYY27 pKa = 8.94HH28 pKa = 6.0NPDD31 pKa = 2.87SGILYY36 pKa = 9.97IAGDD40 pKa = 3.34SGANVGQVSVSGDD53 pKa = 3.34NVVAQIDD60 pKa = 4.1GQSYY64 pKa = 10.56SGASDD69 pKa = 3.87DD70 pKa = 3.63VTDD73 pKa = 5.41IIFIGYY79 pKa = 9.56DD80 pKa = 3.39GNDD83 pKa = 3.19TFTNLSDD90 pKa = 5.01LDD92 pKa = 3.75SSMYY96 pKa = 10.72GHH98 pKa = 7.91LGNDD102 pKa = 3.43TLTGGGGDD110 pKa = 3.95DD111 pKa = 4.02NLVGGAGNDD120 pKa = 3.94TIQGLDD126 pKa = 3.31GDD128 pKa = 4.41DD129 pKa = 4.44RR130 pKa = 11.84IVGADD135 pKa = 3.59GNDD138 pKa = 3.5TLEE141 pKa = 4.81GGDD144 pKa = 3.66GEE146 pKa = 4.98DD147 pKa = 4.95RR148 pKa = 11.84IFGTAGTNTIYY159 pKa = 10.91GGAGDD164 pKa = 3.77DD165 pKa = 3.97TIYY168 pKa = 10.96GSEE171 pKa = 4.51DD172 pKa = 3.21ADD174 pKa = 4.55EE175 pKa = 5.1IYY177 pKa = 11.28GDD179 pKa = 3.66AGADD183 pKa = 3.34KK184 pKa = 10.33IYY186 pKa = 11.1ALGGDD191 pKa = 4.51DD192 pKa = 4.66ILYY195 pKa = 8.98TGTGGVEE202 pKa = 3.7GGSFDD207 pKa = 4.78EE208 pKa = 5.41GDD210 pKa = 3.26LAMGHH215 pKa = 6.34GGDD218 pKa = 4.12DD219 pKa = 3.41EE220 pKa = 6.02FYY222 pKa = 11.1GSTGLNIFYY231 pKa = 10.67GGDD234 pKa = 3.42GDD236 pKa = 5.55DD237 pKa = 3.74IMVGGSGEE245 pKa = 4.08NRR247 pKa = 11.84MHH249 pKa = 6.35GQAGDD254 pKa = 3.62DD255 pKa = 3.7TLTGGEE261 pKa = 4.49KK262 pKa = 10.17GDD264 pKa = 3.91YY265 pKa = 7.46MTGADD270 pKa = 4.9GDD272 pKa = 4.07DD273 pKa = 3.74TFDD276 pKa = 3.32GRR278 pKa = 11.84GGVDD282 pKa = 3.42YY283 pKa = 11.28YY284 pKa = 11.84NVGDD288 pKa = 3.99GSDD291 pKa = 3.73TIVYY295 pKa = 8.42SQSYY299 pKa = 9.08VPTDD303 pKa = 3.05VHH305 pKa = 5.17VTNNGGTAEE314 pKa = 3.98TRR316 pKa = 11.84IQNEE320 pKa = 4.3LVSNATWVQFADD332 pKa = 3.85RR333 pKa = 11.84TISADD338 pKa = 3.0QAYY341 pKa = 10.06YY342 pKa = 9.42LTVDD346 pKa = 3.42EE347 pKa = 5.31TNFSSLNGYY356 pKa = 8.95RR357 pKa = 11.84VSSSVGFVSKK367 pKa = 10.65PSDD370 pKa = 3.51LADD373 pKa = 4.02FAYY376 pKa = 10.45NWSLQMASAGTLSHH390 pKa = 5.8STEE393 pKa = 3.87SDD395 pKa = 3.14RR396 pKa = 11.84ASLYY400 pKa = 10.77VGDD403 pKa = 4.45RR404 pKa = 11.84SAVGEE409 pKa = 4.14NVAMVSDD416 pKa = 4.55TGQSDD421 pKa = 3.96VQIANYY427 pKa = 9.47FLSLWQSSSAHH438 pKa = 5.92NSNLLNSNYY447 pKa = 9.78QEE449 pKa = 3.96VGIGIVKK456 pKa = 10.46SGGAWWATQIFMGG469 pKa = 4.72
Molecular weight: 49.04 kDa
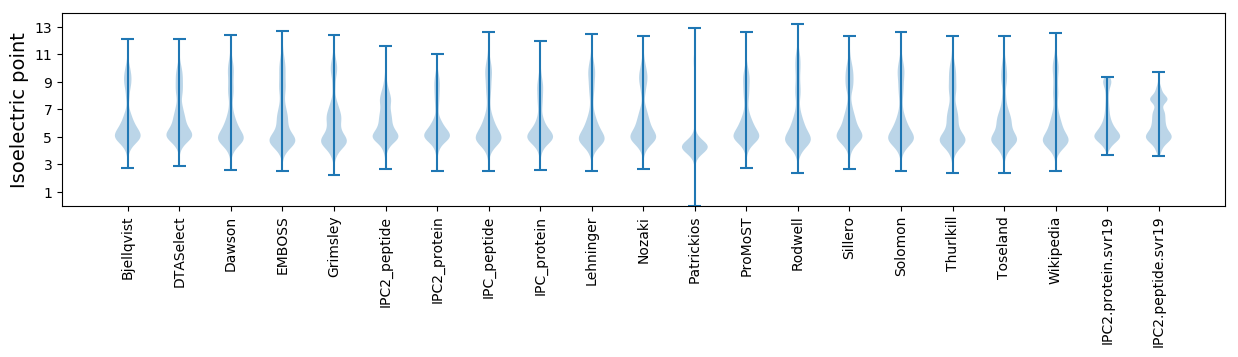
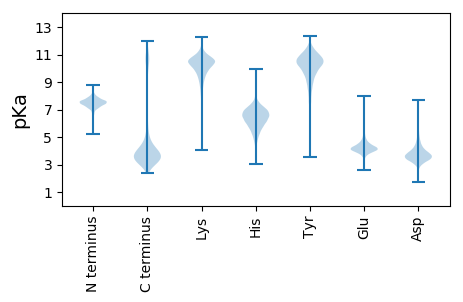
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B9PK37|A0A5B9PK37_9BACT Anhydro-N-acetylmuramic acid kinase OS=Mariniblastus fucicola OX=980251 GN=MFFC18_27390 PE=4 SV=1
MM1 pKa = 7.32NRR3 pKa = 11.84TKK5 pKa = 10.53FFSTIALLCCCFLASPAIAQDD26 pKa = 4.52ISGQWSGGWHH36 pKa = 5.83SGANNHH42 pKa = 6.78RR43 pKa = 11.84GNIKK47 pKa = 9.42ATFCQIDD54 pKa = 4.0SSTMQAKK61 pKa = 9.8FRR63 pKa = 11.84GTFAKK68 pKa = 10.24VIPFRR73 pKa = 11.84YY74 pKa = 5.16TTRR77 pKa = 11.84LNIVSQQPGLTVMAGSRR94 pKa = 11.84RR95 pKa = 11.84LPLGGEE101 pKa = 4.5FKK103 pKa = 10.98YY104 pKa = 10.46QISMTDD110 pKa = 2.81QCFNGTFSSRR120 pKa = 11.84RR121 pKa = 11.84NHH123 pKa = 5.97GQFVMQRR130 pKa = 11.84QQ131 pKa = 3.29
MM1 pKa = 7.32NRR3 pKa = 11.84TKK5 pKa = 10.53FFSTIALLCCCFLASPAIAQDD26 pKa = 4.52ISGQWSGGWHH36 pKa = 5.83SGANNHH42 pKa = 6.78RR43 pKa = 11.84GNIKK47 pKa = 9.42ATFCQIDD54 pKa = 4.0SSTMQAKK61 pKa = 9.8FRR63 pKa = 11.84GTFAKK68 pKa = 10.24VIPFRR73 pKa = 11.84YY74 pKa = 5.16TTRR77 pKa = 11.84LNIVSQQPGLTVMAGSRR94 pKa = 11.84RR95 pKa = 11.84LPLGGEE101 pKa = 4.5FKK103 pKa = 10.98YY104 pKa = 10.46QISMTDD110 pKa = 2.81QCFNGTFSSRR120 pKa = 11.84RR121 pKa = 11.84NHH123 pKa = 5.97GQFVMQRR130 pKa = 11.84QQ131 pKa = 3.29
Molecular weight: 14.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1929523 |
29 |
7782 |
379.6 |
41.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.864 ± 0.033 | 1.122 ± 0.018 |
6.426 ± 0.061 | 6.556 ± 0.035 |
4.262 ± 0.023 | 7.484 ± 0.053 |
1.963 ± 0.02 | 5.545 ± 0.025 |
4.577 ± 0.056 | 9.189 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.361 ± 0.022 | 3.847 ± 0.027 |
4.527 ± 0.025 | 3.913 ± 0.03 |
5.785 ± 0.053 | 7.128 ± 0.034 |
5.494 ± 0.063 | 7.2 ± 0.034 |
1.406 ± 0.017 | 2.354 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
