
STL polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Deltapolyomavirus; Human polyomavirus 11
Average proteome isoelectric point is 5.57
Get precalculated fractions of proteins
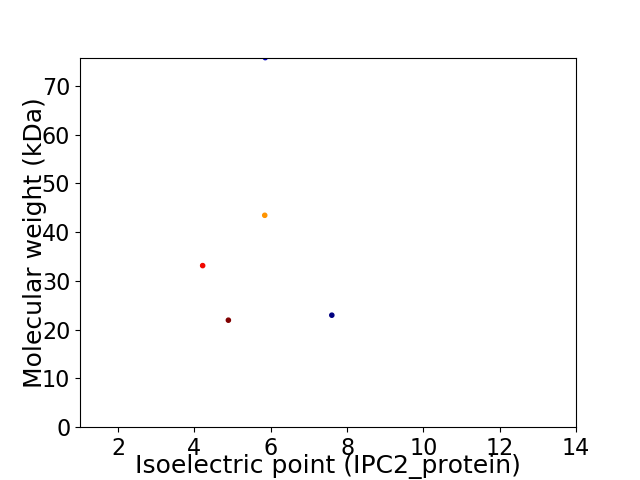
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
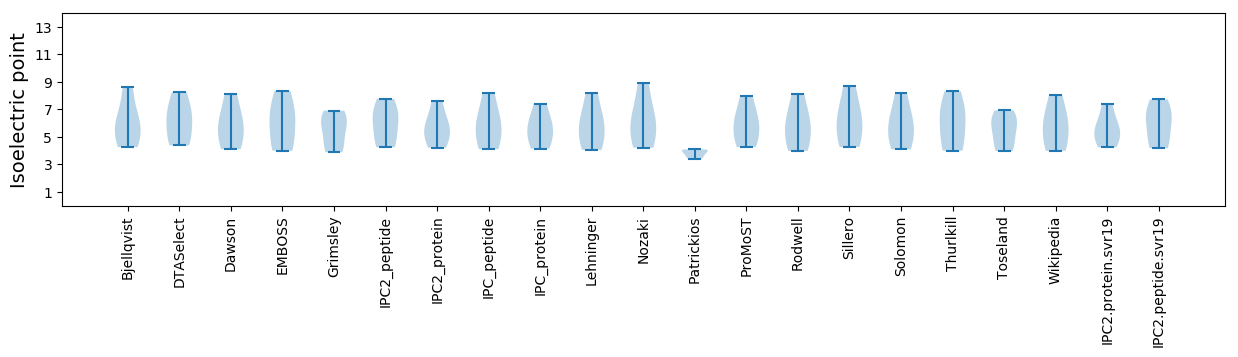
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L7RFL1|L7RFL1_9POLY Small T antigen OS=STL polyomavirus OX=1277649 PE=4 SV=1
MM1 pKa = 7.57GLLISVFLDD10 pKa = 3.43LLDD13 pKa = 4.26LVAITSFDD21 pKa = 3.25VEE23 pKa = 4.88AIVSGDD29 pKa = 3.05AAAIVASQIAAQTVLEE45 pKa = 4.31SAEE48 pKa = 4.05LTEE51 pKa = 4.29VLSSFGLTEE60 pKa = 3.98ASYY63 pKa = 11.0AALSAFPTAFQEE75 pKa = 4.63AVDD78 pKa = 4.43NVLLLQTLSGLSSYY92 pKa = 10.53IAVGLSLQTYY102 pKa = 7.59EE103 pKa = 5.21IPDD106 pKa = 3.67TNMALQIWTPDD117 pKa = 3.08ILDD120 pKa = 4.47EE121 pKa = 4.98IIPGYY126 pKa = 10.33RR127 pKa = 11.84QFQYY131 pKa = 11.38YY132 pKa = 10.8LDD134 pKa = 4.04VLSGWGQSLVRR145 pKa = 11.84TVGRR149 pKa = 11.84AFWEE153 pKa = 4.03AVIDD157 pKa = 3.91EE158 pKa = 4.65TTSRR162 pKa = 11.84LQRR165 pKa = 11.84IGSSSLQAAEE175 pKa = 3.99NAGLQTVEE183 pKa = 4.38NVRR186 pKa = 11.84QAIVTVVEE194 pKa = 3.87NARR197 pKa = 11.84WAIRR201 pKa = 11.84TPVNIYY207 pKa = 10.83GSLSDD212 pKa = 3.91YY213 pKa = 10.83YY214 pKa = 11.58SRR216 pKa = 11.84LPGLNPAQLRR226 pKa = 11.84EE227 pKa = 4.3LNSRR231 pKa = 11.84LQSNAHH237 pKa = 6.44NIQIDD242 pKa = 4.25SYY244 pKa = 10.63SAPDD248 pKa = 3.71SLSGQYY254 pKa = 9.51IEE256 pKa = 5.75RR257 pKa = 11.84PQSYY261 pKa = 9.79SGINMRR267 pKa = 11.84PVPDD271 pKa = 3.48WLLPLILGILGDD283 pKa = 3.92LSPFFEE289 pKa = 4.92EE290 pKa = 3.88IVEE293 pKa = 4.16EE294 pKa = 4.33VEE296 pKa = 3.97NGSKK300 pKa = 10.54KK301 pKa = 10.61KK302 pKa = 10.2II303 pKa = 3.59
MM1 pKa = 7.57GLLISVFLDD10 pKa = 3.43LLDD13 pKa = 4.26LVAITSFDD21 pKa = 3.25VEE23 pKa = 4.88AIVSGDD29 pKa = 3.05AAAIVASQIAAQTVLEE45 pKa = 4.31SAEE48 pKa = 4.05LTEE51 pKa = 4.29VLSSFGLTEE60 pKa = 3.98ASYY63 pKa = 11.0AALSAFPTAFQEE75 pKa = 4.63AVDD78 pKa = 4.43NVLLLQTLSGLSSYY92 pKa = 10.53IAVGLSLQTYY102 pKa = 7.59EE103 pKa = 5.21IPDD106 pKa = 3.67TNMALQIWTPDD117 pKa = 3.08ILDD120 pKa = 4.47EE121 pKa = 4.98IIPGYY126 pKa = 10.33RR127 pKa = 11.84QFQYY131 pKa = 11.38YY132 pKa = 10.8LDD134 pKa = 4.04VLSGWGQSLVRR145 pKa = 11.84TVGRR149 pKa = 11.84AFWEE153 pKa = 4.03AVIDD157 pKa = 3.91EE158 pKa = 4.65TTSRR162 pKa = 11.84LQRR165 pKa = 11.84IGSSSLQAAEE175 pKa = 3.99NAGLQTVEE183 pKa = 4.38NVRR186 pKa = 11.84QAIVTVVEE194 pKa = 3.87NARR197 pKa = 11.84WAIRR201 pKa = 11.84TPVNIYY207 pKa = 10.83GSLSDD212 pKa = 3.91YY213 pKa = 10.83YY214 pKa = 11.58SRR216 pKa = 11.84LPGLNPAQLRR226 pKa = 11.84EE227 pKa = 4.3LNSRR231 pKa = 11.84LQSNAHH237 pKa = 6.44NIQIDD242 pKa = 4.25SYY244 pKa = 10.63SAPDD248 pKa = 3.71SLSGQYY254 pKa = 9.51IEE256 pKa = 5.75RR257 pKa = 11.84PQSYY261 pKa = 9.79SGINMRR267 pKa = 11.84PVPDD271 pKa = 3.48WLLPLILGILGDD283 pKa = 3.92LSPFFEE289 pKa = 4.92EE290 pKa = 3.88IVEE293 pKa = 4.16EE294 pKa = 4.33VEE296 pKa = 3.97NGSKK300 pKa = 10.54KK301 pKa = 10.61KK302 pKa = 10.2II303 pKa = 3.59
Molecular weight: 33.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L7RFL1|L7RFL1_9POLY Small T antigen OS=STL polyomavirus OX=1277649 PE=4 SV=1
MM1 pKa = 7.64DD2 pKa = 3.57QALSRR7 pKa = 11.84AEE9 pKa = 4.1AKK11 pKa = 10.56EE12 pKa = 3.68LMGLLGLPEE21 pKa = 4.96DD22 pKa = 3.61SWGNVPLITYY32 pKa = 8.79RR33 pKa = 11.84FRR35 pKa = 11.84QKK37 pKa = 11.04SKK39 pKa = 10.12IYY41 pKa = 10.69HH42 pKa = 6.42PDD44 pKa = 2.91KK45 pKa = 11.13GGNEE49 pKa = 3.92EE50 pKa = 3.94TMKK53 pKa = 10.87RR54 pKa = 11.84MTEE57 pKa = 4.27LYY59 pKa = 10.96SRR61 pKa = 11.84MQNTLQNLRR70 pKa = 11.84SSNEE74 pKa = 3.87NEE76 pKa = 3.86NMYY79 pKa = 10.56PPVRR83 pKa = 11.84MLLLTDD89 pKa = 4.09TFTLGEE95 pKa = 3.98LLGPQFEE102 pKa = 4.86SKK104 pKa = 10.83VIFIWPTCAKK114 pKa = 9.57CRR116 pKa = 11.84YY117 pKa = 8.27RR118 pKa = 11.84TFCKK122 pKa = 10.18CVCCILKK129 pKa = 10.04RR130 pKa = 11.84QHH132 pKa = 7.26DD133 pKa = 4.51EE134 pKa = 3.73IKK136 pKa = 10.44KK137 pKa = 9.74VRR139 pKa = 11.84NKK141 pKa = 10.56PCVTWGEE148 pKa = 4.84CYY150 pKa = 10.73CFDD153 pKa = 5.77CFLLWFGCDD162 pKa = 3.12LTKK165 pKa = 10.7TSLHH169 pKa = 5.46AWKK172 pKa = 10.02HH173 pKa = 4.04VMYY176 pKa = 10.96NLDD179 pKa = 4.34LDD181 pKa = 4.66LLMFKK186 pKa = 10.21QLNLVSFSFF195 pKa = 3.98
MM1 pKa = 7.64DD2 pKa = 3.57QALSRR7 pKa = 11.84AEE9 pKa = 4.1AKK11 pKa = 10.56EE12 pKa = 3.68LMGLLGLPEE21 pKa = 4.96DD22 pKa = 3.61SWGNVPLITYY32 pKa = 8.79RR33 pKa = 11.84FRR35 pKa = 11.84QKK37 pKa = 11.04SKK39 pKa = 10.12IYY41 pKa = 10.69HH42 pKa = 6.42PDD44 pKa = 2.91KK45 pKa = 11.13GGNEE49 pKa = 3.92EE50 pKa = 3.94TMKK53 pKa = 10.87RR54 pKa = 11.84MTEE57 pKa = 4.27LYY59 pKa = 10.96SRR61 pKa = 11.84MQNTLQNLRR70 pKa = 11.84SSNEE74 pKa = 3.87NEE76 pKa = 3.86NMYY79 pKa = 10.56PPVRR83 pKa = 11.84MLLLTDD89 pKa = 4.09TFTLGEE95 pKa = 3.98LLGPQFEE102 pKa = 4.86SKK104 pKa = 10.83VIFIWPTCAKK114 pKa = 9.57CRR116 pKa = 11.84YY117 pKa = 8.27RR118 pKa = 11.84TFCKK122 pKa = 10.18CVCCILKK129 pKa = 10.04RR130 pKa = 11.84QHH132 pKa = 7.26DD133 pKa = 4.51EE134 pKa = 3.73IKK136 pKa = 10.44KK137 pKa = 9.74VRR139 pKa = 11.84NKK141 pKa = 10.56PCVTWGEE148 pKa = 4.84CYY150 pKa = 10.73CFDD153 pKa = 5.77CFLLWFGCDD162 pKa = 3.12LTKK165 pKa = 10.7TSLHH169 pKa = 5.46AWKK172 pKa = 10.02HH173 pKa = 4.04VMYY176 pKa = 10.96NLDD179 pKa = 4.34LDD181 pKa = 4.66LLMFKK186 pKa = 10.21QLNLVSFSFF195 pKa = 3.98
Molecular weight: 22.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1753 |
195 |
659 |
350.6 |
39.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.362 ± 0.997 | 2.852 ± 0.9 |
5.077 ± 0.207 | 6.275 ± 0.891 |
3.936 ± 0.644 | 5.705 ± 0.632 |
1.483 ± 0.445 | 5.305 ± 1.044 |
4.735 ± 1.419 | 10.667 ± 0.802 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.567 ± 0.571 | 5.476 ± 0.644 |
6.218 ± 1.657 | 4.792 ± 0.455 |
4.963 ± 0.497 | 7.644 ± 0.835 |
6.218 ± 0.967 | 5.819 ± 0.553 |
1.597 ± 0.317 | 3.309 ± 0.375 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
