
Hubei Wuhan insect virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
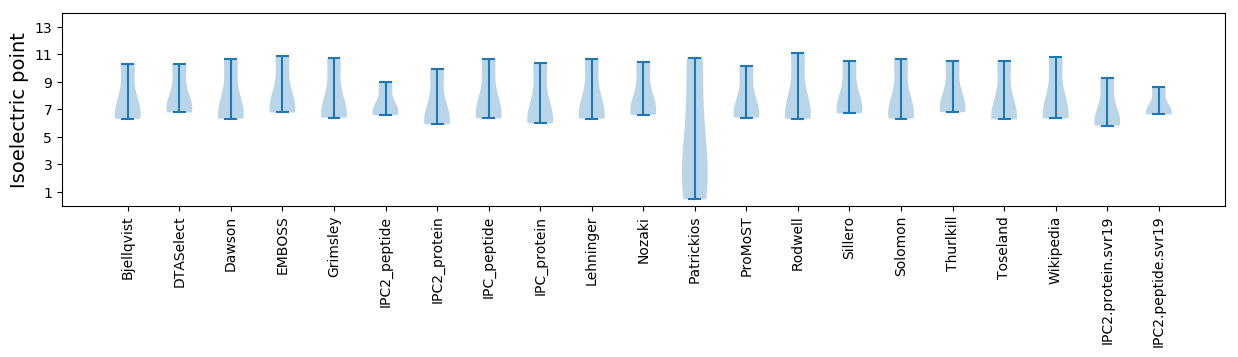
Average proteome isoelectric point is 7.14
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
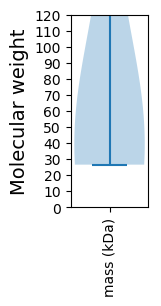
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KK32|A0A1L3KK32_9VIRU RdRp catalytic domain-containing protein OS=Hubei Wuhan insect virus 9 OX=1922833 PE=4 SV=1
MM1 pKa = 7.71DD2 pKa = 3.35VWAKK6 pKa = 10.73AKK8 pKa = 10.35EE9 pKa = 4.09LFDD12 pKa = 3.55KK13 pKa = 10.81LVRR16 pKa = 11.84LVYY19 pKa = 10.72DD20 pKa = 3.87DD21 pKa = 4.62TVHH24 pKa = 7.65DD25 pKa = 5.45DD26 pKa = 3.29ISFTEE31 pKa = 5.16LDD33 pKa = 3.68LQDD36 pKa = 4.51WIDD39 pKa = 3.85TQDD42 pKa = 3.35SNIINLVKK50 pKa = 10.52NIHH53 pKa = 5.92YY54 pKa = 10.59NDD56 pKa = 4.15LKK58 pKa = 10.83VNSYY62 pKa = 11.24NLTIKK67 pKa = 10.62KK68 pKa = 9.77NSKK71 pKa = 9.21PALDD75 pKa = 3.51MSAVNTYY82 pKa = 11.19ASLQTVVFTPKK93 pKa = 10.58DD94 pKa = 3.98FNTLFCPMFKK104 pKa = 9.87IMKK107 pKa = 9.62KK108 pKa = 10.51RR109 pKa = 11.84MIDD112 pKa = 3.25TMNDD116 pKa = 2.85KK117 pKa = 11.07FIMYY121 pKa = 9.75TDD123 pKa = 3.92MPPSKK128 pKa = 10.28FSEE131 pKa = 4.18LLTKK135 pKa = 10.5KK136 pKa = 10.47FSTYY140 pKa = 9.71ILKK143 pKa = 10.09EE144 pKa = 3.9YY145 pKa = 10.47HH146 pKa = 6.63SLEE149 pKa = 4.29FDD151 pKa = 3.28VSKK154 pKa = 11.13YY155 pKa = 10.84DD156 pKa = 3.28KK157 pKa = 10.95SQNLLHH163 pKa = 6.81LLVDD167 pKa = 4.07CMIMRR172 pKa = 11.84HH173 pKa = 5.88FGIPEE178 pKa = 4.14CFVTMWFNGHH188 pKa = 7.12CDD190 pKa = 3.27TKK192 pKa = 11.12LYY194 pKa = 11.08DD195 pKa = 3.89PGNRR199 pKa = 11.84FYY201 pKa = 11.4CDD203 pKa = 2.7VYY205 pKa = 10.35FQRR208 pKa = 11.84KK209 pKa = 9.41SGDD212 pKa = 3.27PSTWLLNTQQILTMIVNCIPDD233 pKa = 3.83SAWAQIILVVASGDD247 pKa = 3.45DD248 pKa = 3.7SEE250 pKa = 5.07IFSTGKK256 pKa = 10.16LQICTSRR263 pKa = 11.84FSNVFNYY270 pKa = 8.63EE271 pKa = 3.7VKK273 pKa = 10.84VYY275 pKa = 11.16DD276 pKa = 4.78KK277 pKa = 9.4FTSMYY282 pKa = 9.21FCSKK286 pKa = 10.19FLIVTEE292 pKa = 4.26YY293 pKa = 8.54GTYY296 pKa = 9.36MLPDD300 pKa = 3.52IYY302 pKa = 11.3KK303 pKa = 10.47LIKK306 pKa = 10.12KK307 pKa = 10.35LGRR310 pKa = 11.84HH311 pKa = 5.48DD312 pKa = 4.28LKK314 pKa = 11.4SRR316 pKa = 11.84DD317 pKa = 3.44HH318 pKa = 6.56VLSFRR323 pKa = 11.84NSVLDD328 pKa = 3.48SLKK331 pKa = 10.9DD332 pKa = 3.43FSVPHH337 pKa = 7.4DD338 pKa = 3.48ILIQYY343 pKa = 7.2EE344 pKa = 3.75KK345 pKa = 11.32SMYY348 pKa = 10.33DD349 pKa = 2.86RR350 pKa = 11.84YY351 pKa = 10.7KK352 pKa = 10.62FPGTISINTVYY363 pKa = 10.68QSLCKK368 pKa = 9.7LTYY371 pKa = 10.37DD372 pKa = 3.8SNAFEE377 pKa = 4.76NLYY380 pKa = 10.29IPNPDD385 pKa = 3.55YY386 pKa = 11.37YY387 pKa = 11.0SNKK390 pKa = 9.58YY391 pKa = 9.99GKK393 pKa = 10.66LSDD396 pKa = 3.7LL397 pKa = 4.28
MM1 pKa = 7.71DD2 pKa = 3.35VWAKK6 pKa = 10.73AKK8 pKa = 10.35EE9 pKa = 4.09LFDD12 pKa = 3.55KK13 pKa = 10.81LVRR16 pKa = 11.84LVYY19 pKa = 10.72DD20 pKa = 3.87DD21 pKa = 4.62TVHH24 pKa = 7.65DD25 pKa = 5.45DD26 pKa = 3.29ISFTEE31 pKa = 5.16LDD33 pKa = 3.68LQDD36 pKa = 4.51WIDD39 pKa = 3.85TQDD42 pKa = 3.35SNIINLVKK50 pKa = 10.52NIHH53 pKa = 5.92YY54 pKa = 10.59NDD56 pKa = 4.15LKK58 pKa = 10.83VNSYY62 pKa = 11.24NLTIKK67 pKa = 10.62KK68 pKa = 9.77NSKK71 pKa = 9.21PALDD75 pKa = 3.51MSAVNTYY82 pKa = 11.19ASLQTVVFTPKK93 pKa = 10.58DD94 pKa = 3.98FNTLFCPMFKK104 pKa = 9.87IMKK107 pKa = 9.62KK108 pKa = 10.51RR109 pKa = 11.84MIDD112 pKa = 3.25TMNDD116 pKa = 2.85KK117 pKa = 11.07FIMYY121 pKa = 9.75TDD123 pKa = 3.92MPPSKK128 pKa = 10.28FSEE131 pKa = 4.18LLTKK135 pKa = 10.5KK136 pKa = 10.47FSTYY140 pKa = 9.71ILKK143 pKa = 10.09EE144 pKa = 3.9YY145 pKa = 10.47HH146 pKa = 6.63SLEE149 pKa = 4.29FDD151 pKa = 3.28VSKK154 pKa = 11.13YY155 pKa = 10.84DD156 pKa = 3.28KK157 pKa = 10.95SQNLLHH163 pKa = 6.81LLVDD167 pKa = 4.07CMIMRR172 pKa = 11.84HH173 pKa = 5.88FGIPEE178 pKa = 4.14CFVTMWFNGHH188 pKa = 7.12CDD190 pKa = 3.27TKK192 pKa = 11.12LYY194 pKa = 11.08DD195 pKa = 3.89PGNRR199 pKa = 11.84FYY201 pKa = 11.4CDD203 pKa = 2.7VYY205 pKa = 10.35FQRR208 pKa = 11.84KK209 pKa = 9.41SGDD212 pKa = 3.27PSTWLLNTQQILTMIVNCIPDD233 pKa = 3.83SAWAQIILVVASGDD247 pKa = 3.45DD248 pKa = 3.7SEE250 pKa = 5.07IFSTGKK256 pKa = 10.16LQICTSRR263 pKa = 11.84FSNVFNYY270 pKa = 8.63EE271 pKa = 3.7VKK273 pKa = 10.84VYY275 pKa = 11.16DD276 pKa = 4.78KK277 pKa = 9.4FTSMYY282 pKa = 9.21FCSKK286 pKa = 10.19FLIVTEE292 pKa = 4.26YY293 pKa = 8.54GTYY296 pKa = 9.36MLPDD300 pKa = 3.52IYY302 pKa = 11.3KK303 pKa = 10.47LIKK306 pKa = 10.12KK307 pKa = 10.35LGRR310 pKa = 11.84HH311 pKa = 5.48DD312 pKa = 4.28LKK314 pKa = 11.4SRR316 pKa = 11.84DD317 pKa = 3.44HH318 pKa = 6.56VLSFRR323 pKa = 11.84NSVLDD328 pKa = 3.48SLKK331 pKa = 10.9DD332 pKa = 3.43FSVPHH337 pKa = 7.4DD338 pKa = 3.48ILIQYY343 pKa = 7.2EE344 pKa = 3.75KK345 pKa = 11.32SMYY348 pKa = 10.33DD349 pKa = 2.86RR350 pKa = 11.84YY351 pKa = 10.7KK352 pKa = 10.62FPGTISINTVYY363 pKa = 10.68QSLCKK368 pKa = 9.7LTYY371 pKa = 10.37DD372 pKa = 3.8SNAFEE377 pKa = 4.76NLYY380 pKa = 10.29IPNPDD385 pKa = 3.55YY386 pKa = 11.37YY387 pKa = 11.0SNKK390 pKa = 9.58YY391 pKa = 9.99GKK393 pKa = 10.66LSDD396 pKa = 3.7LL397 pKa = 4.28
Molecular weight: 46.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KK09|A0A1L3KK09_9VIRU Uncharacterized protein OS=Hubei Wuhan insect virus 9 OX=1922833 PE=4 SV=1
MM1 pKa = 7.77PNTRR5 pKa = 11.84KK6 pKa = 8.02TLVFTNKK13 pKa = 9.56NYY15 pKa = 10.78KK16 pKa = 9.32NNNVNTIQRR25 pKa = 11.84GRR27 pKa = 11.84KK28 pKa = 8.39FNKK31 pKa = 8.54QRR33 pKa = 11.84RR34 pKa = 11.84SRR36 pKa = 11.84SRR38 pKa = 11.84TPRR41 pKa = 11.84RR42 pKa = 11.84SISRR46 pKa = 11.84SRR48 pKa = 11.84SRR50 pKa = 11.84SRR52 pKa = 11.84TSIRR56 pKa = 11.84KK57 pKa = 8.08NIKK60 pKa = 9.4YY61 pKa = 10.52GPIDD65 pKa = 3.92GFLHH69 pKa = 6.94RR70 pKa = 11.84IWSKK74 pKa = 9.31STSLIIDD81 pKa = 4.79PYY83 pKa = 8.9TYY85 pKa = 9.89IGIIIFSILIYY96 pKa = 10.18IHH98 pKa = 6.08QFHH101 pKa = 7.24IDD103 pKa = 3.57DD104 pKa = 3.92SAVNSFIKK112 pKa = 9.46TVKK115 pKa = 10.57KK116 pKa = 10.47NDD118 pKa = 3.59SLKK121 pKa = 10.83PFADD125 pKa = 2.85WFEE128 pKa = 4.38NNINKK133 pKa = 8.82FFALIMVLYY142 pKa = 10.05QSLQISPKK150 pKa = 8.82YY151 pKa = 10.33RR152 pKa = 11.84YY153 pKa = 7.51FTTLISFMLILLLPTLNVATYY174 pKa = 11.35SMALFGSILYY184 pKa = 10.37HH185 pKa = 5.92KK186 pKa = 9.59MKK188 pKa = 10.92YY189 pKa = 10.54KK190 pKa = 9.16MDD192 pKa = 4.75KK193 pKa = 10.46IVILSIFLLAAIWFYY208 pKa = 11.84YY209 pKa = 9.37EE210 pKa = 6.14DD211 pKa = 3.73LMKK214 pKa = 11.01YY215 pKa = 9.57HH216 pKa = 7.01NKK218 pKa = 10.16NINPSVRR225 pKa = 11.84PKK227 pKa = 10.48RR228 pKa = 11.84DD229 pKa = 3.16ADD231 pKa = 3.88DD232 pKa = 4.38VIEE235 pKa = 4.14AA236 pKa = 4.4
MM1 pKa = 7.77PNTRR5 pKa = 11.84KK6 pKa = 8.02TLVFTNKK13 pKa = 9.56NYY15 pKa = 10.78KK16 pKa = 9.32NNNVNTIQRR25 pKa = 11.84GRR27 pKa = 11.84KK28 pKa = 8.39FNKK31 pKa = 8.54QRR33 pKa = 11.84RR34 pKa = 11.84SRR36 pKa = 11.84SRR38 pKa = 11.84TPRR41 pKa = 11.84RR42 pKa = 11.84SISRR46 pKa = 11.84SRR48 pKa = 11.84SRR50 pKa = 11.84SRR52 pKa = 11.84TSIRR56 pKa = 11.84KK57 pKa = 8.08NIKK60 pKa = 9.4YY61 pKa = 10.52GPIDD65 pKa = 3.92GFLHH69 pKa = 6.94RR70 pKa = 11.84IWSKK74 pKa = 9.31STSLIIDD81 pKa = 4.79PYY83 pKa = 8.9TYY85 pKa = 9.89IGIIIFSILIYY96 pKa = 10.18IHH98 pKa = 6.08QFHH101 pKa = 7.24IDD103 pKa = 3.57DD104 pKa = 3.92SAVNSFIKK112 pKa = 9.46TVKK115 pKa = 10.57KK116 pKa = 10.47NDD118 pKa = 3.59SLKK121 pKa = 10.83PFADD125 pKa = 2.85WFEE128 pKa = 4.38NNINKK133 pKa = 8.82FFALIMVLYY142 pKa = 10.05QSLQISPKK150 pKa = 8.82YY151 pKa = 10.33RR152 pKa = 11.84YY153 pKa = 7.51FTTLISFMLILLLPTLNVATYY174 pKa = 11.35SMALFGSILYY184 pKa = 10.37HH185 pKa = 5.92KK186 pKa = 9.59MKK188 pKa = 10.92YY189 pKa = 10.54KK190 pKa = 9.16MDD192 pKa = 4.75KK193 pKa = 10.46IVILSIFLLAAIWFYY208 pKa = 11.84YY209 pKa = 9.37EE210 pKa = 6.14DD211 pKa = 3.73LMKK214 pKa = 11.01YY215 pKa = 9.57HH216 pKa = 7.01NKK218 pKa = 10.16NINPSVRR225 pKa = 11.84PKK227 pKa = 10.48RR228 pKa = 11.84DD229 pKa = 3.16ADD231 pKa = 3.88DD232 pKa = 4.38VIEE235 pKa = 4.14AA236 pKa = 4.4
Molecular weight: 27.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4076 |
224 |
2418 |
679.3 |
79.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.999 ± 0.362 | 1.742 ± 0.337 |
7.139 ± 0.906 | 3.729 ± 0.681 |
5.397 ± 0.79 | 2.92 ± 0.297 |
2.87 ± 0.199 | 8.415 ± 1.051 |
8.194 ± 0.519 | 7.728 ± 0.348 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.582 ± 0.112 | 5.618 ± 0.218 |
3.263 ± 0.327 | 2.355 ± 0.097 |
3.606 ± 0.75 | 8.023 ± 0.477 |
7.041 ± 0.454 | 6.722 ± 0.543 |
0.711 ± 0.148 | 6.943 ± 0.428 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
