
Apis mellifera associated microvirus 48
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.72
Get precalculated fractions of proteins
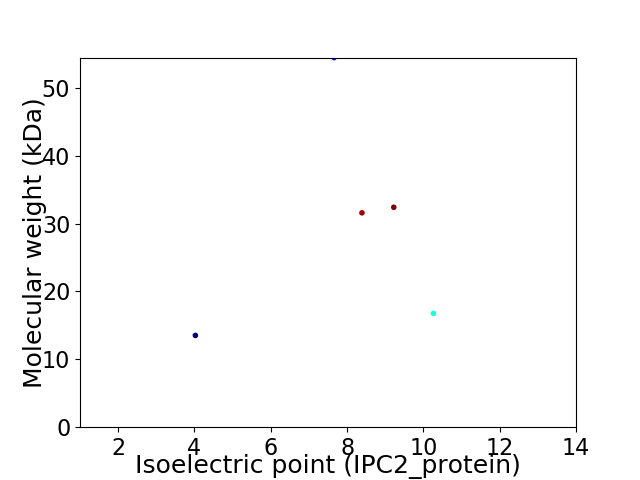
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UTP4|A0A3S8UTP4_9VIRU Uncharacterized protein OS=Apis mellifera associated microvirus 48 OX=2494778 PE=4 SV=1
MM1 pKa = 7.77EE2 pKa = 4.27FTMIKK7 pKa = 8.12DD8 pKa = 3.15TSMIYY13 pKa = 8.36QTLDD17 pKa = 3.28DD18 pKa = 4.99TPVSMPTRR26 pKa = 11.84LRR28 pKa = 11.84LPQSRR33 pKa = 11.84TEE35 pKa = 3.9QIRR38 pKa = 11.84QYY40 pKa = 10.81VRR42 pKa = 11.84EE43 pKa = 4.08EE44 pKa = 3.82MSRR47 pKa = 11.84AAAEE51 pKa = 3.95QGHH54 pKa = 5.99EE55 pKa = 4.33SFEE58 pKa = 4.21EE59 pKa = 4.73ADD61 pKa = 4.89DD62 pKa = 4.42FSLDD66 pKa = 4.23DD67 pKa = 4.56EE68 pKa = 4.97EE69 pKa = 6.33DD70 pKa = 3.83MPLSPYY76 pKa = 9.53EE77 pKa = 4.04LAMLEE82 pKa = 4.31PTPPLQNGVSAEE94 pKa = 4.07GAPPVGPAATSEE106 pKa = 4.4VPAASQPVTSEE117 pKa = 4.04TPPNVPP123 pKa = 3.94
MM1 pKa = 7.77EE2 pKa = 4.27FTMIKK7 pKa = 8.12DD8 pKa = 3.15TSMIYY13 pKa = 8.36QTLDD17 pKa = 3.28DD18 pKa = 4.99TPVSMPTRR26 pKa = 11.84LRR28 pKa = 11.84LPQSRR33 pKa = 11.84TEE35 pKa = 3.9QIRR38 pKa = 11.84QYY40 pKa = 10.81VRR42 pKa = 11.84EE43 pKa = 4.08EE44 pKa = 3.82MSRR47 pKa = 11.84AAAEE51 pKa = 3.95QGHH54 pKa = 5.99EE55 pKa = 4.33SFEE58 pKa = 4.21EE59 pKa = 4.73ADD61 pKa = 4.89DD62 pKa = 4.42FSLDD66 pKa = 4.23DD67 pKa = 4.56EE68 pKa = 4.97EE69 pKa = 6.33DD70 pKa = 3.83MPLSPYY76 pKa = 9.53EE77 pKa = 4.04LAMLEE82 pKa = 4.31PTPPLQNGVSAEE94 pKa = 4.07GAPPVGPAATSEE106 pKa = 4.4VPAASQPVTSEE117 pKa = 4.04TPPNVPP123 pKa = 3.94
Molecular weight: 13.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q8U4V2|A0A3Q8U4V2_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 48 OX=2494778 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.09CIKK5 pKa = 10.34LATRR9 pKa = 11.84TDD11 pKa = 3.75GFQFPCGQCKK21 pKa = 9.38NCRR24 pKa = 11.84INHH27 pKa = 5.14RR28 pKa = 11.84RR29 pKa = 11.84DD30 pKa = 2.98WQSRR34 pKa = 11.84LLLEE38 pKa = 4.8ASSHH42 pKa = 4.9AHH44 pKa = 5.23SCFVTLTFADD54 pKa = 3.65TGIPPILRR62 pKa = 11.84RR63 pKa = 11.84SDD65 pKa = 3.13LKK67 pKa = 11.0RR68 pKa = 11.84FFRR71 pKa = 11.84ALRR74 pKa = 11.84VYY76 pKa = 10.51IPDD79 pKa = 3.42ARR81 pKa = 11.84HH82 pKa = 5.54FSVGEE87 pKa = 3.6YY88 pKa = 10.34GKK90 pKa = 10.64RR91 pKa = 11.84FGRR94 pKa = 11.84AHH96 pKa = 5.63YY97 pKa = 7.39HH98 pKa = 3.77THH100 pKa = 6.14IFSNVPVLATHH111 pKa = 7.59LAACWPFGHH120 pKa = 6.72VDD122 pKa = 3.26IGDD125 pKa = 3.93TEE127 pKa = 4.28PASLDD132 pKa = 3.4YY133 pKa = 10.99TLGYY137 pKa = 9.76LLKK140 pKa = 10.54DD141 pKa = 3.17KK142 pKa = 10.78RR143 pKa = 11.84SNVWPIEE150 pKa = 3.77SRR152 pKa = 11.84FPEE155 pKa = 4.13FRR157 pKa = 11.84AFSPGIGKK165 pKa = 8.41MALPHH170 pKa = 6.51LLIDD174 pKa = 4.22GNLLPRR180 pKa = 11.84EE181 pKa = 4.0YY182 pKa = 10.87KK183 pKa = 10.81VFGNLWPVSRR193 pKa = 11.84YY194 pKa = 9.23LRR196 pKa = 11.84QRR198 pKa = 11.84AKK200 pKa = 11.15KK201 pKa = 9.19MGFDD205 pKa = 3.58VSQTEE210 pKa = 4.2EE211 pKa = 3.77KK212 pKa = 10.52RR213 pKa = 11.84LEE215 pKa = 4.09EE216 pKa = 4.81FEE218 pKa = 4.21AQQMRR223 pKa = 11.84AVLRR227 pKa = 11.84NPSLTQAEE235 pKa = 4.65VSTLYY240 pKa = 11.01DD241 pKa = 3.24GFVKK245 pKa = 10.27KK246 pKa = 10.32RR247 pKa = 11.84QEE249 pKa = 3.82KK250 pKa = 11.14LEE252 pKa = 3.96VLKK255 pKa = 11.07KK256 pKa = 10.23KK257 pKa = 10.57AIRR260 pKa = 11.84DD261 pKa = 3.73AYY263 pKa = 9.74RR264 pKa = 11.84QIHH267 pKa = 5.14GHH269 pKa = 4.91VKK271 pKa = 8.87TRR273 pKa = 11.84SKK275 pKa = 11.53NEE277 pKa = 3.83TFF279 pKa = 3.71
MM1 pKa = 7.5KK2 pKa = 10.09CIKK5 pKa = 10.34LATRR9 pKa = 11.84TDD11 pKa = 3.75GFQFPCGQCKK21 pKa = 9.38NCRR24 pKa = 11.84INHH27 pKa = 5.14RR28 pKa = 11.84RR29 pKa = 11.84DD30 pKa = 2.98WQSRR34 pKa = 11.84LLLEE38 pKa = 4.8ASSHH42 pKa = 4.9AHH44 pKa = 5.23SCFVTLTFADD54 pKa = 3.65TGIPPILRR62 pKa = 11.84RR63 pKa = 11.84SDD65 pKa = 3.13LKK67 pKa = 11.0RR68 pKa = 11.84FFRR71 pKa = 11.84ALRR74 pKa = 11.84VYY76 pKa = 10.51IPDD79 pKa = 3.42ARR81 pKa = 11.84HH82 pKa = 5.54FSVGEE87 pKa = 3.6YY88 pKa = 10.34GKK90 pKa = 10.64RR91 pKa = 11.84FGRR94 pKa = 11.84AHH96 pKa = 5.63YY97 pKa = 7.39HH98 pKa = 3.77THH100 pKa = 6.14IFSNVPVLATHH111 pKa = 7.59LAACWPFGHH120 pKa = 6.72VDD122 pKa = 3.26IGDD125 pKa = 3.93TEE127 pKa = 4.28PASLDD132 pKa = 3.4YY133 pKa = 10.99TLGYY137 pKa = 9.76LLKK140 pKa = 10.54DD141 pKa = 3.17KK142 pKa = 10.78RR143 pKa = 11.84SNVWPIEE150 pKa = 3.77SRR152 pKa = 11.84FPEE155 pKa = 4.13FRR157 pKa = 11.84AFSPGIGKK165 pKa = 8.41MALPHH170 pKa = 6.51LLIDD174 pKa = 4.22GNLLPRR180 pKa = 11.84EE181 pKa = 4.0YY182 pKa = 10.87KK183 pKa = 10.81VFGNLWPVSRR193 pKa = 11.84YY194 pKa = 9.23LRR196 pKa = 11.84QRR198 pKa = 11.84AKK200 pKa = 11.15KK201 pKa = 9.19MGFDD205 pKa = 3.58VSQTEE210 pKa = 4.2EE211 pKa = 3.77KK212 pKa = 10.52RR213 pKa = 11.84LEE215 pKa = 4.09EE216 pKa = 4.81FEE218 pKa = 4.21AQQMRR223 pKa = 11.84AVLRR227 pKa = 11.84NPSLTQAEE235 pKa = 4.65VSTLYY240 pKa = 11.01DD241 pKa = 3.24GFVKK245 pKa = 10.27KK246 pKa = 10.32RR247 pKa = 11.84QEE249 pKa = 3.82KK250 pKa = 11.14LEE252 pKa = 3.96VLKK255 pKa = 11.07KK256 pKa = 10.23KK257 pKa = 10.57AIRR260 pKa = 11.84DD261 pKa = 3.73AYY263 pKa = 9.74RR264 pKa = 11.84QIHH267 pKa = 5.14GHH269 pKa = 4.91VKK271 pKa = 8.87TRR273 pKa = 11.84SKK275 pKa = 11.53NEE277 pKa = 3.83TFF279 pKa = 3.71
Molecular weight: 32.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1344 |
123 |
495 |
268.8 |
29.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.896 ± 0.92 | 0.744 ± 0.397 |
4.911 ± 0.227 | 5.134 ± 1.082 |
4.464 ± 0.547 | 7.292 ± 0.997 |
2.307 ± 0.581 | 3.646 ± 0.216 |
4.241 ± 0.891 | 8.408 ± 0.417 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.753 ± 0.439 | 2.679 ± 0.494 |
6.771 ± 1.096 | 4.167 ± 0.407 |
8.185 ± 1.101 | 7.813 ± 0.892 |
6.92 ± 0.967 | 5.655 ± 0.397 |
1.042 ± 0.289 | 2.976 ± 0.498 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
