
Streptomyces sp. SCSIO 3032
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
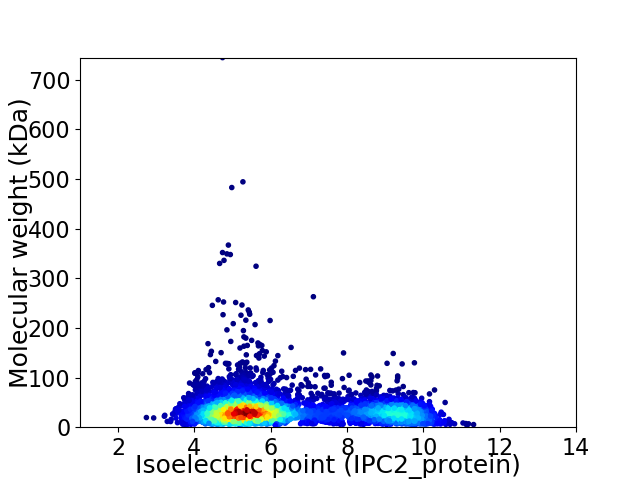
Virtual 2D-PAGE plot for 5248 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W7CZK4|A0A1W7CZK4_9ACTN SAM-dependent methyltransferase OS=Streptomyces sp. SCSIO 3032 OX=1264596 GN=CAG99_16560 PE=4 SV=1
MM1 pKa = 7.24SQPPPPPQNPPGPPQPEE18 pKa = 4.29PAAGPAPAPGGPAPASGGPAPASGGPASGTPAPAPGGPAPASGGPASGTPAPAPGGPAPASGGPASGTPAPAPSGPAPASGGPASGTPAPAPGTPASAPPAVPGGLPGQPGYY130 pKa = 11.02GYY132 pKa = 8.74PQQAYY137 pKa = 9.77GSPQQGYY144 pKa = 8.57GFPQSPGAAPYY155 pKa = 8.65PSPYY159 pKa = 9.04QQPGQPGPTAPGGAFGAPTAPGPYY183 pKa = 9.99GPATGGFATPPPGPGGPKK201 pKa = 9.2GRR203 pKa = 11.84GRR205 pKa = 11.84RR206 pKa = 11.84GVVLAAVAAFAVLVGGGGAVWLLTGVQDD234 pKa = 5.25DD235 pKa = 4.3PAPAAGDD242 pKa = 3.93GADD245 pKa = 3.87GGADD249 pKa = 3.53DD250 pKa = 4.72GAEE253 pKa = 3.99GGARR257 pKa = 11.84GEE259 pKa = 4.33AGNGAGLPLPTEE271 pKa = 4.88AIGASLAWEE280 pKa = 4.53VPAPEE285 pKa = 4.0VSEE288 pKa = 4.55DD289 pKa = 3.74EE290 pKa = 4.6NIIDD294 pKa = 5.95AKK296 pKa = 9.47GTWLIEE302 pKa = 4.03DD303 pKa = 3.48AFVRR307 pKa = 11.84VTPDD311 pKa = 2.87SLVSYY316 pKa = 10.69DD317 pKa = 4.05LATGQEE323 pKa = 4.13NWSLPFEE330 pKa = 4.35FSGGDD335 pKa = 3.67CLPSSNVSDD344 pKa = 3.45NRR346 pKa = 11.84VALLQGRR353 pKa = 11.84DD354 pKa = 3.84CEE356 pKa = 4.43VLTVVDD362 pKa = 4.25IAAGTEE368 pKa = 4.19VMSMQIEE375 pKa = 4.68SEE377 pKa = 4.22WPTSAGSVPAILGDD391 pKa = 3.78TVAVGTGVAGLGFSISRR408 pKa = 11.84QEE410 pKa = 4.31MIWRR414 pKa = 11.84PPANSDD420 pKa = 3.87CRR422 pKa = 11.84EE423 pKa = 3.75SDD425 pKa = 3.86YY426 pKa = 11.61LVVDD430 pKa = 4.34DD431 pKa = 5.2MFVSQLSCGFAGDD444 pKa = 3.53EE445 pKa = 4.47GGSIRR450 pKa = 11.84ATDD453 pKa = 3.48EE454 pKa = 4.46SGDD457 pKa = 3.72TEE459 pKa = 4.19LWEE462 pKa = 3.96WAYY465 pKa = 10.69GPTYY469 pKa = 9.96EE470 pKa = 4.81GEE472 pKa = 4.11PFEE475 pKa = 4.37VQSVVSVDD483 pKa = 3.66PLVVTAAVGEE493 pKa = 4.76DD494 pKa = 3.72YY495 pKa = 10.19TAADD499 pKa = 3.75HH500 pKa = 7.19ARR502 pKa = 11.84VLVIDD507 pKa = 4.2EE508 pKa = 4.28DD509 pKa = 4.19HH510 pKa = 6.91EE511 pKa = 4.47EE512 pKa = 4.19VAVEE516 pKa = 4.7LDD518 pKa = 3.55YY519 pKa = 11.66DD520 pKa = 3.49QDD522 pKa = 3.94RR523 pKa = 11.84YY524 pKa = 10.18MRR526 pKa = 11.84PCEE529 pKa = 3.94TNSFDD534 pKa = 3.94NCWLGVEE541 pKa = 4.64HH542 pKa = 7.3EE543 pKa = 4.69GFLYY547 pKa = 10.73LPSNVPYY554 pKa = 10.73GDD556 pKa = 3.7NAVEE560 pKa = 4.86AFDD563 pKa = 5.59LSTGQPLWEE572 pKa = 4.11VPAINGGQILPFGVQDD588 pKa = 4.07GQILAYY594 pKa = 9.88QVASTEE600 pKa = 4.1LEE602 pKa = 4.17GMVVAIDD609 pKa = 3.64PATEE613 pKa = 3.84EE614 pKa = 4.05VRR616 pKa = 11.84PIMALDD622 pKa = 3.75RR623 pKa = 11.84AARR626 pKa = 11.84PQEE629 pKa = 3.94HH630 pKa = 6.82ALMSGLFVHH639 pKa = 6.94DD640 pKa = 3.14QHH642 pKa = 8.66IIWRR646 pKa = 11.84DD647 pKa = 3.31NTLVLANRR655 pKa = 11.84SFYY658 pKa = 11.21SSDD661 pKa = 3.58GEE663 pKa = 4.23DD664 pKa = 3.25VRR666 pKa = 11.84SVLVYY671 pKa = 10.38RR672 pKa = 4.98
MM1 pKa = 7.24SQPPPPPQNPPGPPQPEE18 pKa = 4.29PAAGPAPAPGGPAPASGGPAPASGGPASGTPAPAPGGPAPASGGPASGTPAPAPGGPAPASGGPASGTPAPAPSGPAPASGGPASGTPAPAPGTPASAPPAVPGGLPGQPGYY130 pKa = 11.02GYY132 pKa = 8.74PQQAYY137 pKa = 9.77GSPQQGYY144 pKa = 8.57GFPQSPGAAPYY155 pKa = 8.65PSPYY159 pKa = 9.04QQPGQPGPTAPGGAFGAPTAPGPYY183 pKa = 9.99GPATGGFATPPPGPGGPKK201 pKa = 9.2GRR203 pKa = 11.84GRR205 pKa = 11.84RR206 pKa = 11.84GVVLAAVAAFAVLVGGGGAVWLLTGVQDD234 pKa = 5.25DD235 pKa = 4.3PAPAAGDD242 pKa = 3.93GADD245 pKa = 3.87GGADD249 pKa = 3.53DD250 pKa = 4.72GAEE253 pKa = 3.99GGARR257 pKa = 11.84GEE259 pKa = 4.33AGNGAGLPLPTEE271 pKa = 4.88AIGASLAWEE280 pKa = 4.53VPAPEE285 pKa = 4.0VSEE288 pKa = 4.55DD289 pKa = 3.74EE290 pKa = 4.6NIIDD294 pKa = 5.95AKK296 pKa = 9.47GTWLIEE302 pKa = 4.03DD303 pKa = 3.48AFVRR307 pKa = 11.84VTPDD311 pKa = 2.87SLVSYY316 pKa = 10.69DD317 pKa = 4.05LATGQEE323 pKa = 4.13NWSLPFEE330 pKa = 4.35FSGGDD335 pKa = 3.67CLPSSNVSDD344 pKa = 3.45NRR346 pKa = 11.84VALLQGRR353 pKa = 11.84DD354 pKa = 3.84CEE356 pKa = 4.43VLTVVDD362 pKa = 4.25IAAGTEE368 pKa = 4.19VMSMQIEE375 pKa = 4.68SEE377 pKa = 4.22WPTSAGSVPAILGDD391 pKa = 3.78TVAVGTGVAGLGFSISRR408 pKa = 11.84QEE410 pKa = 4.31MIWRR414 pKa = 11.84PPANSDD420 pKa = 3.87CRR422 pKa = 11.84EE423 pKa = 3.75SDD425 pKa = 3.86YY426 pKa = 11.61LVVDD430 pKa = 4.34DD431 pKa = 5.2MFVSQLSCGFAGDD444 pKa = 3.53EE445 pKa = 4.47GGSIRR450 pKa = 11.84ATDD453 pKa = 3.48EE454 pKa = 4.46SGDD457 pKa = 3.72TEE459 pKa = 4.19LWEE462 pKa = 3.96WAYY465 pKa = 10.69GPTYY469 pKa = 9.96EE470 pKa = 4.81GEE472 pKa = 4.11PFEE475 pKa = 4.37VQSVVSVDD483 pKa = 3.66PLVVTAAVGEE493 pKa = 4.76DD494 pKa = 3.72YY495 pKa = 10.19TAADD499 pKa = 3.75HH500 pKa = 7.19ARR502 pKa = 11.84VLVIDD507 pKa = 4.2EE508 pKa = 4.28DD509 pKa = 4.19HH510 pKa = 6.91EE511 pKa = 4.47EE512 pKa = 4.19VAVEE516 pKa = 4.7LDD518 pKa = 3.55YY519 pKa = 11.66DD520 pKa = 3.49QDD522 pKa = 3.94RR523 pKa = 11.84YY524 pKa = 10.18MRR526 pKa = 11.84PCEE529 pKa = 3.94TNSFDD534 pKa = 3.94NCWLGVEE541 pKa = 4.64HH542 pKa = 7.3EE543 pKa = 4.69GFLYY547 pKa = 10.73LPSNVPYY554 pKa = 10.73GDD556 pKa = 3.7NAVEE560 pKa = 4.86AFDD563 pKa = 5.59LSTGQPLWEE572 pKa = 4.11VPAINGGQILPFGVQDD588 pKa = 4.07GQILAYY594 pKa = 9.88QVASTEE600 pKa = 4.1LEE602 pKa = 4.17GMVVAIDD609 pKa = 3.64PATEE613 pKa = 3.84EE614 pKa = 4.05VRR616 pKa = 11.84PIMALDD622 pKa = 3.75RR623 pKa = 11.84AARR626 pKa = 11.84PQEE629 pKa = 3.94HH630 pKa = 6.82ALMSGLFVHH639 pKa = 6.94DD640 pKa = 3.14QHH642 pKa = 8.66IIWRR646 pKa = 11.84DD647 pKa = 3.31NTLVLANRR655 pKa = 11.84SFYY658 pKa = 11.21SSDD661 pKa = 3.58GEE663 pKa = 4.23DD664 pKa = 3.25VRR666 pKa = 11.84SVLVYY671 pKa = 10.38RR672 pKa = 4.98
Molecular weight: 67.83 kDa
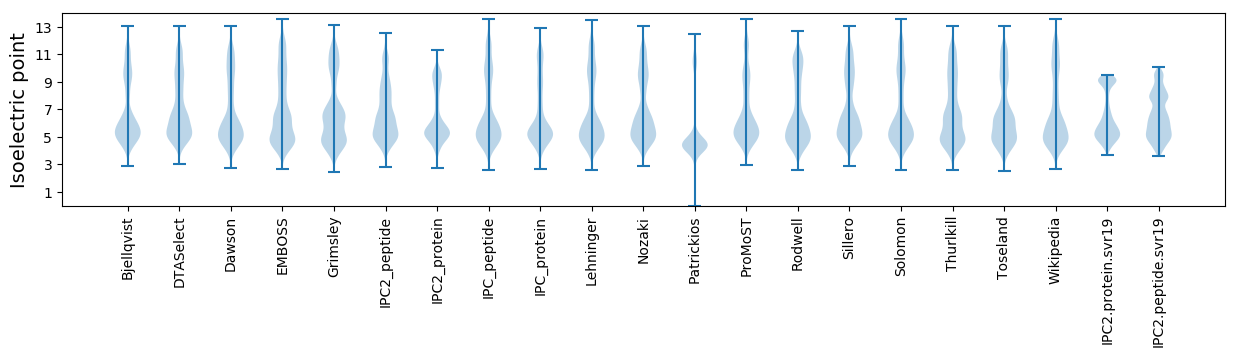
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W7CYE1|A0A1W7CYE1_9ACTN Uncharacterized protein OS=Streptomyces sp. SCSIO 3032 OX=1264596 GN=CAG99_13450 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.82GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.82GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1788051 |
29 |
7193 |
340.7 |
36.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.667 ± 0.06 | 0.75 ± 0.009 |
6.052 ± 0.029 | 5.953 ± 0.032 |
2.658 ± 0.018 | 9.97 ± 0.042 |
2.373 ± 0.017 | 2.828 ± 0.022 |
1.335 ± 0.019 | 10.408 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.613 ± 0.015 | 1.58 ± 0.019 |
6.463 ± 0.037 | 2.435 ± 0.02 |
8.897 ± 0.045 | 4.515 ± 0.024 |
5.777 ± 0.031 | 8.341 ± 0.033 |
1.521 ± 0.014 | 1.862 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
