
Oscillibacter valericigenes (strain DSM 18026 / NBRC 101213 / Sjm18-20)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Oscillibacter; Oscillibacter valericigenes
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
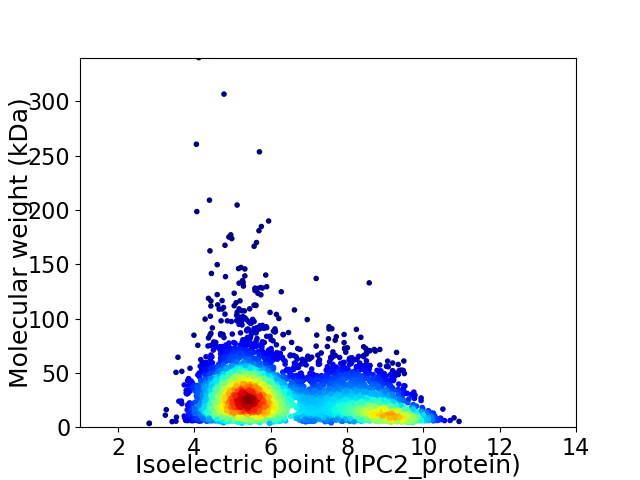
Virtual 2D-PAGE plot for 4593 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G4KRV1|G4KRV1_OSCVS Putative transposase orfB for insertion sequence element OS=Oscillibacter valericigenes (strain DSM 18026 / NBRC 101213 / Sjm18-20) OX=693746 GN=OBV_27690 PE=4 SV=1
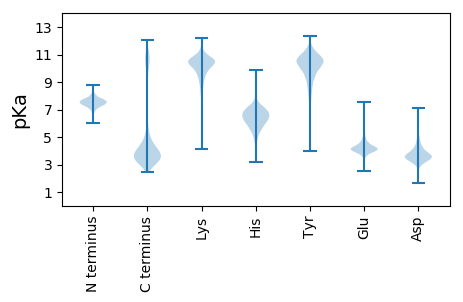
MM1 pKa = 7.28KK2 pKa = 10.26RR3 pKa = 11.84AFLAALAAMLLLSGCGKK20 pKa = 9.75QAEE23 pKa = 4.64EE24 pKa = 4.13PSGLYY29 pKa = 10.24YY30 pKa = 10.16EE31 pKa = 5.08ASGIAPDD38 pKa = 4.12TVLLTVDD45 pKa = 3.36GRR47 pKa = 11.84EE48 pKa = 4.19VPAARR53 pKa = 11.84YY54 pKa = 7.94FYY56 pKa = 10.46WLTADD61 pKa = 3.89CDD63 pKa = 4.04YY64 pKa = 11.18LAEE67 pKa = 4.16QSGGEE72 pKa = 4.03PDD74 pKa = 3.12WTGDD78 pKa = 4.06AGGQTLDD85 pKa = 4.13EE86 pKa = 4.32YY87 pKa = 11.73VKK89 pKa = 11.01AQALSAAVFYY99 pKa = 9.3ATLEE103 pKa = 4.21SLAEE107 pKa = 4.05SHH109 pKa = 7.5DD110 pKa = 4.25CVLTAGDD117 pKa = 4.5LAAMDD122 pKa = 5.19KK123 pKa = 11.0DD124 pKa = 3.69WADD127 pKa = 3.28QAAAAGGEE135 pKa = 4.04DD136 pKa = 4.0AYY138 pKa = 11.48LAVLAQKK145 pKa = 11.01GLDD148 pKa = 3.35QAGAQLFSADD158 pKa = 3.56YY159 pKa = 10.67YY160 pKa = 11.18LYY162 pKa = 10.17EE163 pKa = 4.05QLRR166 pKa = 11.84EE167 pKa = 4.01LSQTEE172 pKa = 4.36GNDD175 pKa = 3.6LAPKK179 pKa = 10.3SGDD182 pKa = 3.14VTAYY186 pKa = 9.99AQAAGLYY193 pKa = 8.73TADD196 pKa = 3.8TLLFSDD202 pKa = 4.43GDD204 pKa = 3.98LAAVALSEE212 pKa = 4.77LEE214 pKa = 4.37KK215 pKa = 11.33DD216 pKa = 3.68PDD218 pKa = 4.31LDD220 pKa = 4.18PATLKK225 pKa = 10.99DD226 pKa = 3.36GTYY229 pKa = 10.9ASVTAAPGDD238 pKa = 3.74GTLPEE243 pKa = 4.29AAEE246 pKa = 4.22SALKK250 pKa = 10.64GLQTDD255 pKa = 3.22QWSQPVEE262 pKa = 4.35TEE264 pKa = 3.53AGVYY268 pKa = 10.07LLRR271 pKa = 11.84PAALDD276 pKa = 3.31EE277 pKa = 4.85DD278 pKa = 4.37AAGDD282 pKa = 3.51LWFDD286 pKa = 4.63GRR288 pKa = 11.84LQDD291 pKa = 4.63LAADD295 pKa = 4.06APVTLAEE302 pKa = 4.5EE303 pKa = 4.52YY304 pKa = 11.22GSFTAASFYY313 pKa = 10.92KK314 pKa = 9.91GLCAARR320 pKa = 11.84VGGKK324 pKa = 9.43
MM1 pKa = 7.28KK2 pKa = 10.26RR3 pKa = 11.84AFLAALAAMLLLSGCGKK20 pKa = 9.75QAEE23 pKa = 4.64EE24 pKa = 4.13PSGLYY29 pKa = 10.24YY30 pKa = 10.16EE31 pKa = 5.08ASGIAPDD38 pKa = 4.12TVLLTVDD45 pKa = 3.36GRR47 pKa = 11.84EE48 pKa = 4.19VPAARR53 pKa = 11.84YY54 pKa = 7.94FYY56 pKa = 10.46WLTADD61 pKa = 3.89CDD63 pKa = 4.04YY64 pKa = 11.18LAEE67 pKa = 4.16QSGGEE72 pKa = 4.03PDD74 pKa = 3.12WTGDD78 pKa = 4.06AGGQTLDD85 pKa = 4.13EE86 pKa = 4.32YY87 pKa = 11.73VKK89 pKa = 11.01AQALSAAVFYY99 pKa = 9.3ATLEE103 pKa = 4.21SLAEE107 pKa = 4.05SHH109 pKa = 7.5DD110 pKa = 4.25CVLTAGDD117 pKa = 4.5LAAMDD122 pKa = 5.19KK123 pKa = 11.0DD124 pKa = 3.69WADD127 pKa = 3.28QAAAAGGEE135 pKa = 4.04DD136 pKa = 4.0AYY138 pKa = 11.48LAVLAQKK145 pKa = 11.01GLDD148 pKa = 3.35QAGAQLFSADD158 pKa = 3.56YY159 pKa = 10.67YY160 pKa = 11.18LYY162 pKa = 10.17EE163 pKa = 4.05QLRR166 pKa = 11.84EE167 pKa = 4.01LSQTEE172 pKa = 4.36GNDD175 pKa = 3.6LAPKK179 pKa = 10.3SGDD182 pKa = 3.14VTAYY186 pKa = 9.99AQAAGLYY193 pKa = 8.73TADD196 pKa = 3.8TLLFSDD202 pKa = 4.43GDD204 pKa = 3.98LAAVALSEE212 pKa = 4.77LEE214 pKa = 4.37KK215 pKa = 11.33DD216 pKa = 3.68PDD218 pKa = 4.31LDD220 pKa = 4.18PATLKK225 pKa = 10.99DD226 pKa = 3.36GTYY229 pKa = 10.9ASVTAAPGDD238 pKa = 3.74GTLPEE243 pKa = 4.29AAEE246 pKa = 4.22SALKK250 pKa = 10.64GLQTDD255 pKa = 3.22QWSQPVEE262 pKa = 4.35TEE264 pKa = 3.53AGVYY268 pKa = 10.07LLRR271 pKa = 11.84PAALDD276 pKa = 3.31EE277 pKa = 4.85DD278 pKa = 4.37AAGDD282 pKa = 3.51LWFDD286 pKa = 4.63GRR288 pKa = 11.84LQDD291 pKa = 4.63LAADD295 pKa = 4.06APVTLAEE302 pKa = 4.5EE303 pKa = 4.52YY304 pKa = 11.22GSFTAASFYY313 pKa = 10.92KK314 pKa = 9.91GLCAARR320 pKa = 11.84VGGKK324 pKa = 9.43
Molecular weight: 33.96 kDa
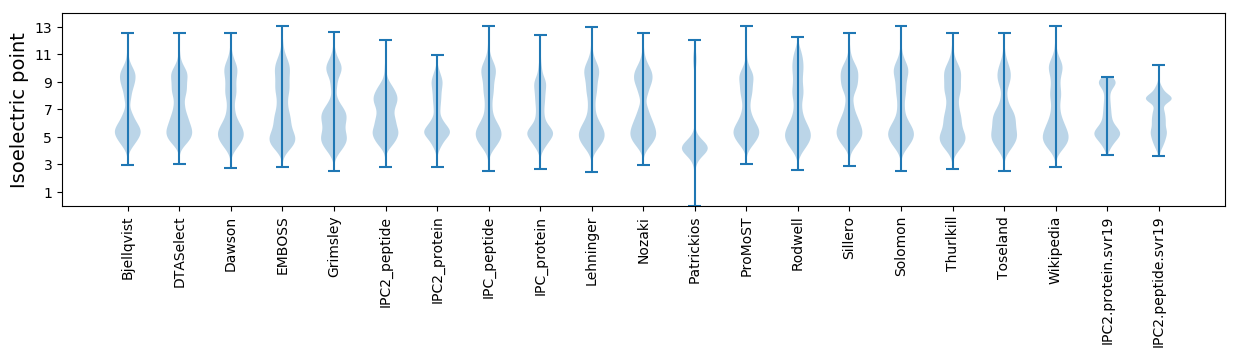
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G4KZ25|G4KZ25_OSCVS Stage 0 sporulation protein A homolog OS=Oscillibacter valericigenes (strain DSM 18026 / NBRC 101213 / Sjm18-20) OX=693746 GN=OBV_40180 PE=4 SV=1
MM1 pKa = 7.86AGRR4 pKa = 11.84NTQLPLARR12 pKa = 11.84MRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84TMSPVLRR23 pKa = 11.84SRR25 pKa = 11.84LTGLLSRR32 pKa = 11.84RR33 pKa = 11.84NTASHH38 pKa = 6.65RR39 pKa = 11.84CIFSCFRR46 pKa = 11.84RR47 pKa = 11.84GTRR50 pKa = 11.84ALPRR54 pKa = 11.84LPFPAWPGAVSAISSPQSKK73 pKa = 9.82IKK75 pKa = 10.82AGAA78 pKa = 3.24
MM1 pKa = 7.86AGRR4 pKa = 11.84NTQLPLARR12 pKa = 11.84MRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84TMSPVLRR23 pKa = 11.84SRR25 pKa = 11.84LTGLLSRR32 pKa = 11.84RR33 pKa = 11.84NTASHH38 pKa = 6.65RR39 pKa = 11.84CIFSCFRR46 pKa = 11.84RR47 pKa = 11.84GTRR50 pKa = 11.84ALPRR54 pKa = 11.84LPFPAWPGAVSAISSPQSKK73 pKa = 9.82IKK75 pKa = 10.82AGAA78 pKa = 3.24
Molecular weight: 8.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1245691 |
31 |
3221 |
271.2 |
29.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.254 ± 0.048 | 1.719 ± 0.018 |
5.503 ± 0.035 | 6.408 ± 0.043 |
3.94 ± 0.027 | 7.643 ± 0.038 |
1.789 ± 0.016 | 5.939 ± 0.04 |
5.379 ± 0.035 | 9.608 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.842 ± 0.022 | 3.737 ± 0.026 |
3.943 ± 0.027 | 3.418 ± 0.025 |
5.447 ± 0.041 | 6.18 ± 0.042 |
5.763 ± 0.044 | 6.933 ± 0.034 |
1.015 ± 0.013 | 3.54 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
