
Oceanobacillus piezotolerans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Oceanobacillus
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
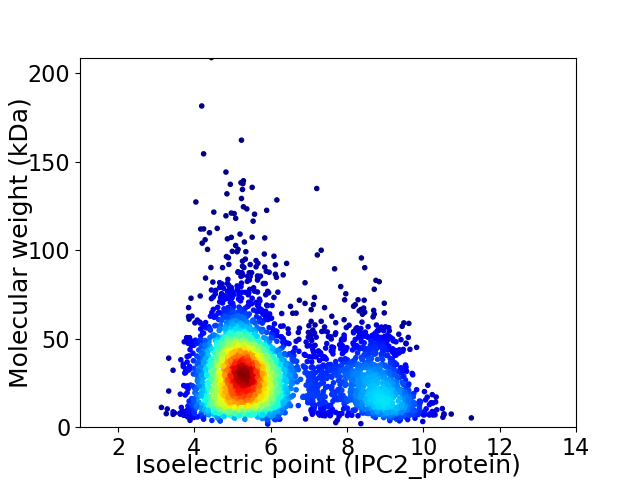
Virtual 2D-PAGE plot for 3815 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
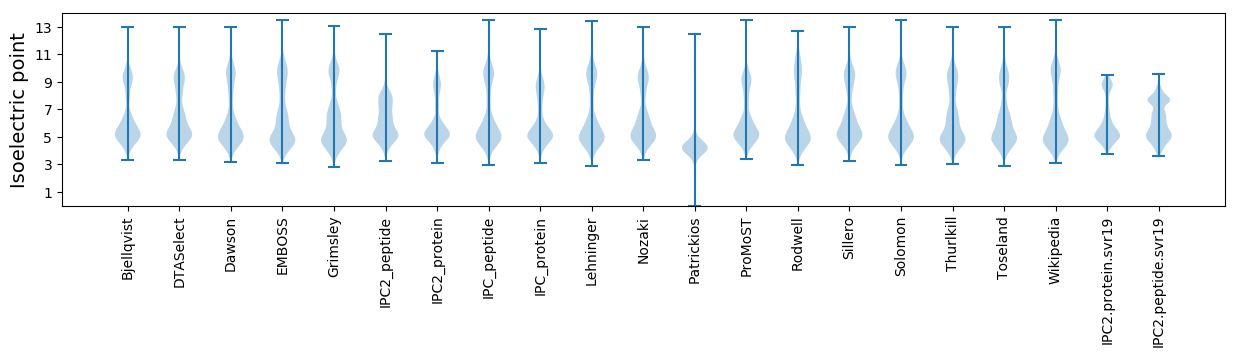
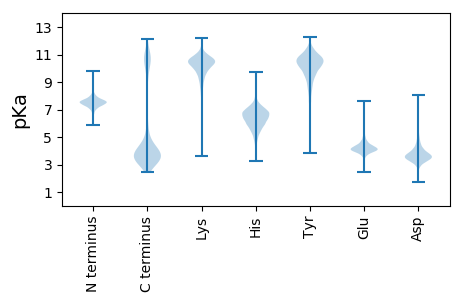
Protein with the lowest isoelectric point:
>tr|A0A498D8X5|A0A498D8X5_9BACI DUF3231 family protein OS=Oceanobacillus piezotolerans OX=2448030 GN=D8M04_14425 PE=4 SV=1
MM1 pKa = 6.94GTSFLYY7 pKa = 10.13HH8 pKa = 4.69YY9 pKa = 10.09HH10 pKa = 6.25CLKK13 pKa = 10.18NRR15 pKa = 11.84RR16 pKa = 11.84GFDD19 pKa = 3.51EE20 pKa = 4.79VILRR24 pKa = 11.84NWKK27 pKa = 8.64WRR29 pKa = 11.84ITALISMGLVLLLAACGSGEE49 pKa = 4.1EE50 pKa = 4.49TSSEE54 pKa = 4.04EE55 pKa = 4.43NGASDD60 pKa = 4.26AAEE63 pKa = 3.91YY64 pKa = 10.92DD65 pKa = 3.94LNDD68 pKa = 3.53SYY70 pKa = 11.74TVVSDD75 pKa = 3.39NSFVPFEE82 pKa = 4.0FVEE85 pKa = 4.16NGEE88 pKa = 4.08LQGFDD93 pKa = 3.38IEE95 pKa = 4.87IINAIAEE102 pKa = 4.16EE103 pKa = 4.36AGFEE107 pKa = 4.12IAGGEE112 pKa = 4.03IEE114 pKa = 4.35TTNFDD119 pKa = 4.04GIIPGLQTGQFEE131 pKa = 4.13IAIAGISITEE141 pKa = 4.19DD142 pKa = 2.9RR143 pKa = 11.84AKK145 pKa = 10.94VIDD148 pKa = 4.03YY149 pKa = 10.44SDD151 pKa = 3.97PYY153 pKa = 9.58YY154 pKa = 10.83EE155 pKa = 4.07SGITIGVPIDD165 pKa = 3.64NEE167 pKa = 4.67DD168 pKa = 3.51IQSLEE173 pKa = 4.01DD174 pKa = 3.83LEE176 pKa = 4.84GKK178 pKa = 7.98TVATRR183 pKa = 11.84LGSTSANFIRR193 pKa = 11.84EE194 pKa = 3.99NLDD197 pKa = 3.69DD198 pKa = 3.86VTLNEE203 pKa = 4.25YY204 pKa = 9.81EE205 pKa = 4.4QLDD208 pKa = 3.93QVYY211 pKa = 9.83LAVEE215 pKa = 4.25NGSADD220 pKa = 3.32AVVYY224 pKa = 8.63DD225 pKa = 4.36APNVNYY231 pKa = 10.45YY232 pKa = 10.17ISTTGEE238 pKa = 4.18GKK240 pKa = 10.47LKK242 pKa = 10.88SIDD245 pKa = 3.75EE246 pKa = 4.64LYY248 pKa = 10.4QAEE251 pKa = 4.74DD252 pKa = 3.4YY253 pKa = 11.34GIAITKK259 pKa = 10.15GNEE262 pKa = 4.07DD263 pKa = 4.04LVSAINDD270 pKa = 3.43ALATLKK276 pKa = 10.99EE277 pKa = 4.03NGTYY281 pKa = 10.72DD282 pKa = 3.75EE283 pKa = 5.79IYY285 pKa = 10.38EE286 pKa = 4.22KK287 pKa = 10.53WFGAAEE293 pKa = 4.02EE294 pKa = 4.26
MM1 pKa = 6.94GTSFLYY7 pKa = 10.13HH8 pKa = 4.69YY9 pKa = 10.09HH10 pKa = 6.25CLKK13 pKa = 10.18NRR15 pKa = 11.84RR16 pKa = 11.84GFDD19 pKa = 3.51EE20 pKa = 4.79VILRR24 pKa = 11.84NWKK27 pKa = 8.64WRR29 pKa = 11.84ITALISMGLVLLLAACGSGEE49 pKa = 4.1EE50 pKa = 4.49TSSEE54 pKa = 4.04EE55 pKa = 4.43NGASDD60 pKa = 4.26AAEE63 pKa = 3.91YY64 pKa = 10.92DD65 pKa = 3.94LNDD68 pKa = 3.53SYY70 pKa = 11.74TVVSDD75 pKa = 3.39NSFVPFEE82 pKa = 4.0FVEE85 pKa = 4.16NGEE88 pKa = 4.08LQGFDD93 pKa = 3.38IEE95 pKa = 4.87IINAIAEE102 pKa = 4.16EE103 pKa = 4.36AGFEE107 pKa = 4.12IAGGEE112 pKa = 4.03IEE114 pKa = 4.35TTNFDD119 pKa = 4.04GIIPGLQTGQFEE131 pKa = 4.13IAIAGISITEE141 pKa = 4.19DD142 pKa = 2.9RR143 pKa = 11.84AKK145 pKa = 10.94VIDD148 pKa = 4.03YY149 pKa = 10.44SDD151 pKa = 3.97PYY153 pKa = 9.58YY154 pKa = 10.83EE155 pKa = 4.07SGITIGVPIDD165 pKa = 3.64NEE167 pKa = 4.67DD168 pKa = 3.51IQSLEE173 pKa = 4.01DD174 pKa = 3.83LEE176 pKa = 4.84GKK178 pKa = 7.98TVATRR183 pKa = 11.84LGSTSANFIRR193 pKa = 11.84EE194 pKa = 3.99NLDD197 pKa = 3.69DD198 pKa = 3.86VTLNEE203 pKa = 4.25YY204 pKa = 9.81EE205 pKa = 4.4QLDD208 pKa = 3.93QVYY211 pKa = 9.83LAVEE215 pKa = 4.25NGSADD220 pKa = 3.32AVVYY224 pKa = 8.63DD225 pKa = 4.36APNVNYY231 pKa = 10.45YY232 pKa = 10.17ISTTGEE238 pKa = 4.18GKK240 pKa = 10.47LKK242 pKa = 10.88SIDD245 pKa = 3.75EE246 pKa = 4.64LYY248 pKa = 10.4QAEE251 pKa = 4.74DD252 pKa = 3.4YY253 pKa = 11.34GIAITKK259 pKa = 10.15GNEE262 pKa = 4.07DD263 pKa = 4.04LVSAINDD270 pKa = 3.43ALATLKK276 pKa = 10.99EE277 pKa = 4.03NGTYY281 pKa = 10.72DD282 pKa = 3.75EE283 pKa = 5.79IYY285 pKa = 10.38EE286 pKa = 4.22KK287 pKa = 10.53WFGAAEE293 pKa = 4.02EE294 pKa = 4.26
Molecular weight: 32.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A498D2Z8|A0A498D2Z8_9BACI SRPBCC family protein OS=Oceanobacillus piezotolerans OX=2448030 GN=D8M04_15830 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.22KK14 pKa = 8.69VHH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.48VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.22KK14 pKa = 8.69VHH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.48VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1080255 |
15 |
1849 |
283.2 |
31.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.835 ± 0.042 | 0.601 ± 0.011 |
5.172 ± 0.035 | 7.885 ± 0.048 |
4.516 ± 0.033 | 6.836 ± 0.038 |
2.065 ± 0.022 | 8.353 ± 0.035 |
6.701 ± 0.038 | 9.616 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.761 ± 0.02 | 4.711 ± 0.025 |
3.562 ± 0.024 | 3.613 ± 0.024 |
3.918 ± 0.026 | 5.928 ± 0.032 |
5.389 ± 0.024 | 6.918 ± 0.035 |
0.996 ± 0.014 | 3.624 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
