
Kazachstania africana (strain ATCC 22294 / BCRC 22015 / CBS 2517 / CECT 1963 / NBRC 1671 / NRRL Y-8276) (Yeast) (Kluyveromyces africanus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetaceae; Kazachstania; Kazachstania africana
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
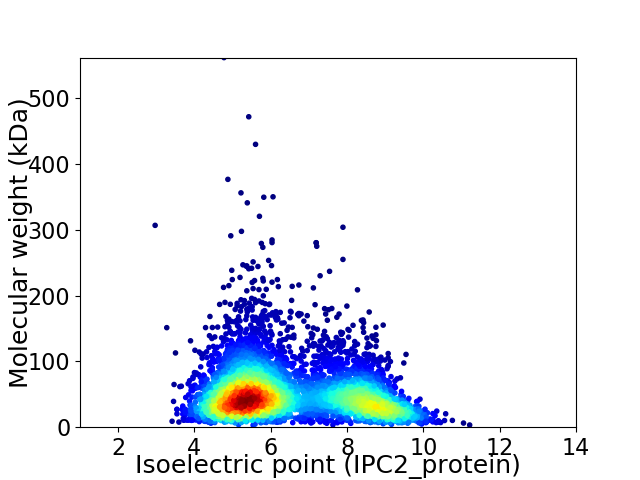
Virtual 2D-PAGE plot for 5354 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H2AS48|H2AS48_KAZAF Uncharacterized protein OS=Kazachstania africana (strain ATCC 22294 / BCRC 22015 / CBS 2517 / CECT 1963 / NBRC 1671 / NRRL Y-8276) OX=1071382 GN=KAFR0C02050 PE=4 SV=1
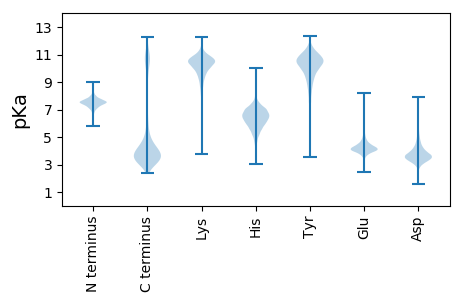
MM1 pKa = 7.3QLQDD5 pKa = 4.56LLLSLSVAISTAHH18 pKa = 6.2AWSPTDD24 pKa = 3.13SYY26 pKa = 12.13APGEE30 pKa = 4.31ITCDD34 pKa = 3.15ADD36 pKa = 3.6VNLVRR41 pKa = 11.84NATDD45 pKa = 3.66LSSDD49 pKa = 3.42EE50 pKa = 4.21QEE52 pKa = 3.96WLKK55 pKa = 11.11KK56 pKa = 9.96RR57 pKa = 11.84DD58 pKa = 3.99TITEE62 pKa = 4.09SAMKK66 pKa = 9.93TFLTRR71 pKa = 11.84ATANFSDD78 pKa = 3.79TSFLDD83 pKa = 3.55SLFEE87 pKa = 4.09NSSYY91 pKa = 10.67VPRR94 pKa = 11.84VGIAASGGGYY104 pKa = 9.96RR105 pKa = 11.84AMLSGAGMLAAFDD118 pKa = 3.62NRR120 pKa = 11.84TRR122 pKa = 11.84GANEE126 pKa = 3.71NGLGGLLQATTYY138 pKa = 11.11LSGLSGGNWMTGTLAWNNWTSVQEE162 pKa = 4.46IIDD165 pKa = 4.14SFSDD169 pKa = 3.72SNTSSIWDD177 pKa = 3.38ITNSIVTPGGNNSTFTSEE195 pKa = 3.77RR196 pKa = 11.84FASIISDD203 pKa = 3.15IDD205 pKa = 3.69DD206 pKa = 3.73KK207 pKa = 11.81QEE209 pKa = 3.77AGFDD213 pKa = 3.33ITLVDD218 pKa = 2.95VWGRR222 pKa = 11.84MLSYY226 pKa = 11.06YY227 pKa = 9.51FFPSLPNGGAAYY239 pKa = 8.0TWSTLRR245 pKa = 11.84DD246 pKa = 2.92ADD248 pKa = 3.54VFMNAEE254 pKa = 4.15MPFPISVADD263 pKa = 3.81GQYY266 pKa = 10.4PGTSVDD272 pKa = 3.88SLNSTVFEE280 pKa = 4.22FNPFEE285 pKa = 4.28MGSWDD290 pKa = 3.35PTLNAFTDD298 pKa = 3.73VKK300 pKa = 11.23YY301 pKa = 10.95LGTNVTNGSPIEE313 pKa = 4.17SGKK316 pKa = 10.77CIAGFDD322 pKa = 3.59NVGFIIGTSSDD333 pKa = 3.17ILNDD337 pKa = 2.99ITSSYY342 pKa = 10.42EE343 pKa = 3.66AVLTYY348 pKa = 10.93LEE350 pKa = 4.61TEE352 pKa = 4.31FLSDD356 pKa = 3.24STATSDD362 pKa = 3.45EE363 pKa = 4.41YY364 pKa = 11.48TLGIYY369 pKa = 10.17SPNPFKK375 pKa = 10.78GTTNIDD381 pKa = 3.55SNYY384 pKa = 8.02STSIVDD390 pKa = 3.56SDD392 pKa = 4.06SLYY395 pKa = 11.02LADD398 pKa = 4.44GGEE401 pKa = 4.5DD402 pKa = 3.88GEE404 pKa = 4.71VVPFIPLIKK413 pKa = 9.83KK414 pKa = 8.9EE415 pKa = 4.04RR416 pKa = 11.84EE417 pKa = 3.54IDD419 pKa = 3.65VVFAFDD425 pKa = 4.77NDD427 pKa = 4.3DD428 pKa = 5.73GSDD431 pKa = 3.65DD432 pKa = 3.86WPSGSNLISTYY443 pKa = 9.28EE444 pKa = 4.0RR445 pKa = 11.84QFSSQGKK452 pKa = 9.39SIAFPYY458 pKa = 10.59VPDD461 pKa = 4.22LATFSDD467 pKa = 3.7LGLNEE472 pKa = 4.14RR473 pKa = 11.84PTFFGCDD480 pKa = 3.14ASNLTDD486 pKa = 4.16LAYY489 pKa = 10.38VPPLIVYY496 pKa = 9.49IPNAQYY502 pKa = 10.89SYY504 pKa = 10.94PSNTSTYY511 pKa = 7.89TLSYY515 pKa = 11.09SMEE518 pKa = 3.94EE519 pKa = 3.87RR520 pKa = 11.84LEE522 pKa = 4.23MIQNGYY528 pKa = 8.97EE529 pKa = 3.61AATRR533 pKa = 11.84NNLTEE538 pKa = 4.36DD539 pKa = 3.77SGFLGCVGCAVIRR552 pKa = 11.84RR553 pKa = 11.84KK554 pKa = 9.83QEE556 pKa = 3.83SLNITLPSEE565 pKa = 4.5CEE567 pKa = 3.63QCFANYY573 pKa = 9.07CWNGTLTDD581 pKa = 3.71VSSVSSSNSSYY592 pKa = 11.61SNSTTTLSSGAAEE605 pKa = 4.2STTASSNASNSSSTSSSSAASSSSAAKK632 pKa = 10.35ANAAASVEE640 pKa = 4.23TGKK643 pKa = 10.77LFSLLTAFGVLFSLMM658 pKa = 3.86
MM1 pKa = 7.3QLQDD5 pKa = 4.56LLLSLSVAISTAHH18 pKa = 6.2AWSPTDD24 pKa = 3.13SYY26 pKa = 12.13APGEE30 pKa = 4.31ITCDD34 pKa = 3.15ADD36 pKa = 3.6VNLVRR41 pKa = 11.84NATDD45 pKa = 3.66LSSDD49 pKa = 3.42EE50 pKa = 4.21QEE52 pKa = 3.96WLKK55 pKa = 11.11KK56 pKa = 9.96RR57 pKa = 11.84DD58 pKa = 3.99TITEE62 pKa = 4.09SAMKK66 pKa = 9.93TFLTRR71 pKa = 11.84ATANFSDD78 pKa = 3.79TSFLDD83 pKa = 3.55SLFEE87 pKa = 4.09NSSYY91 pKa = 10.67VPRR94 pKa = 11.84VGIAASGGGYY104 pKa = 9.96RR105 pKa = 11.84AMLSGAGMLAAFDD118 pKa = 3.62NRR120 pKa = 11.84TRR122 pKa = 11.84GANEE126 pKa = 3.71NGLGGLLQATTYY138 pKa = 11.11LSGLSGGNWMTGTLAWNNWTSVQEE162 pKa = 4.46IIDD165 pKa = 4.14SFSDD169 pKa = 3.72SNTSSIWDD177 pKa = 3.38ITNSIVTPGGNNSTFTSEE195 pKa = 3.77RR196 pKa = 11.84FASIISDD203 pKa = 3.15IDD205 pKa = 3.69DD206 pKa = 3.73KK207 pKa = 11.81QEE209 pKa = 3.77AGFDD213 pKa = 3.33ITLVDD218 pKa = 2.95VWGRR222 pKa = 11.84MLSYY226 pKa = 11.06YY227 pKa = 9.51FFPSLPNGGAAYY239 pKa = 8.0TWSTLRR245 pKa = 11.84DD246 pKa = 2.92ADD248 pKa = 3.54VFMNAEE254 pKa = 4.15MPFPISVADD263 pKa = 3.81GQYY266 pKa = 10.4PGTSVDD272 pKa = 3.88SLNSTVFEE280 pKa = 4.22FNPFEE285 pKa = 4.28MGSWDD290 pKa = 3.35PTLNAFTDD298 pKa = 3.73VKK300 pKa = 11.23YY301 pKa = 10.95LGTNVTNGSPIEE313 pKa = 4.17SGKK316 pKa = 10.77CIAGFDD322 pKa = 3.59NVGFIIGTSSDD333 pKa = 3.17ILNDD337 pKa = 2.99ITSSYY342 pKa = 10.42EE343 pKa = 3.66AVLTYY348 pKa = 10.93LEE350 pKa = 4.61TEE352 pKa = 4.31FLSDD356 pKa = 3.24STATSDD362 pKa = 3.45EE363 pKa = 4.41YY364 pKa = 11.48TLGIYY369 pKa = 10.17SPNPFKK375 pKa = 10.78GTTNIDD381 pKa = 3.55SNYY384 pKa = 8.02STSIVDD390 pKa = 3.56SDD392 pKa = 4.06SLYY395 pKa = 11.02LADD398 pKa = 4.44GGEE401 pKa = 4.5DD402 pKa = 3.88GEE404 pKa = 4.71VVPFIPLIKK413 pKa = 9.83KK414 pKa = 8.9EE415 pKa = 4.04RR416 pKa = 11.84EE417 pKa = 3.54IDD419 pKa = 3.65VVFAFDD425 pKa = 4.77NDD427 pKa = 4.3DD428 pKa = 5.73GSDD431 pKa = 3.65DD432 pKa = 3.86WPSGSNLISTYY443 pKa = 9.28EE444 pKa = 4.0RR445 pKa = 11.84QFSSQGKK452 pKa = 9.39SIAFPYY458 pKa = 10.59VPDD461 pKa = 4.22LATFSDD467 pKa = 3.7LGLNEE472 pKa = 4.14RR473 pKa = 11.84PTFFGCDD480 pKa = 3.14ASNLTDD486 pKa = 4.16LAYY489 pKa = 10.38VPPLIVYY496 pKa = 9.49IPNAQYY502 pKa = 10.89SYY504 pKa = 10.94PSNTSTYY511 pKa = 7.89TLSYY515 pKa = 11.09SMEE518 pKa = 3.94EE519 pKa = 3.87RR520 pKa = 11.84LEE522 pKa = 4.23MIQNGYY528 pKa = 8.97EE529 pKa = 3.61AATRR533 pKa = 11.84NNLTEE538 pKa = 4.36DD539 pKa = 3.77SGFLGCVGCAVIRR552 pKa = 11.84RR553 pKa = 11.84KK554 pKa = 9.83QEE556 pKa = 3.83SLNITLPSEE565 pKa = 4.5CEE567 pKa = 3.63QCFANYY573 pKa = 9.07CWNGTLTDD581 pKa = 3.71VSSVSSSNSSYY592 pKa = 11.61SNSTTTLSSGAAEE605 pKa = 4.2STTASSNASNSSSTSSSSAASSSSAAKK632 pKa = 10.35ANAAASVEE640 pKa = 4.23TGKK643 pKa = 10.77LFSLLTAFGVLFSLMM658 pKa = 3.86
Molecular weight: 70.66 kDa
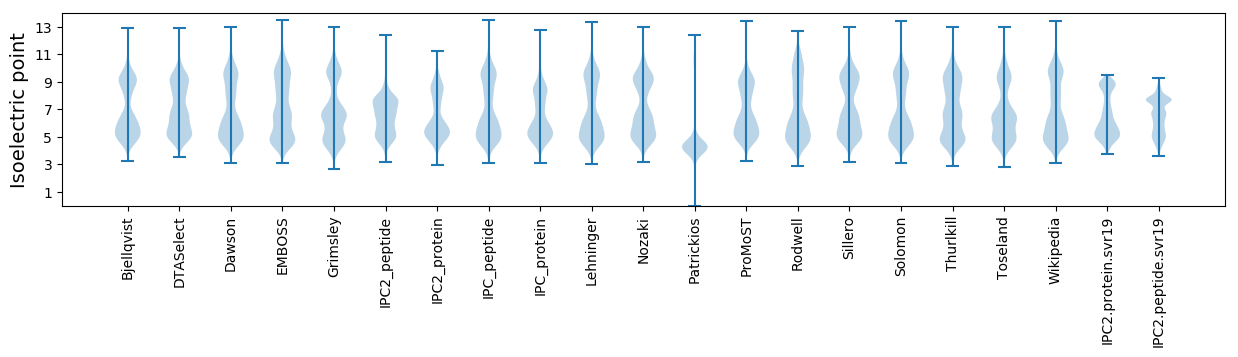
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H2B2C9|H2B2C9_KAZAF SEC7 domain-containing protein OS=Kazachstania africana (strain ATCC 22294 / BCRC 22015 / CBS 2517 / CECT 1963 / NBRC 1671 / NRRL Y-8276) OX=1071382 GN=KAFR0L01690 PE=4 SV=1
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.09WRR6 pKa = 11.84KK7 pKa = 8.87KK8 pKa = 8.73RR9 pKa = 11.84VRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.71RR15 pKa = 11.84KK16 pKa = 8.27RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.19VRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 10.98
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.09WRR6 pKa = 11.84KK7 pKa = 8.87KK8 pKa = 8.73RR9 pKa = 11.84VRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.71RR15 pKa = 11.84KK16 pKa = 8.27RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.19VRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 10.98
Molecular weight: 3.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2609361 |
25 |
4933 |
487.4 |
55.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.193 ± 0.024 | 1.18 ± 0.011 |
5.985 ± 0.022 | 6.705 ± 0.037 |
4.554 ± 0.019 | 4.698 ± 0.03 |
2.037 ± 0.012 | 6.994 ± 0.025 |
7.409 ± 0.034 | 9.724 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.061 ± 0.01 | 6.577 ± 0.032 |
3.993 ± 0.02 | 3.761 ± 0.021 |
4.273 ± 0.021 | 8.912 ± 0.049 |
5.952 ± 0.033 | 5.525 ± 0.023 |
1.008 ± 0.01 | 3.459 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
