
Haloactinobacterium album
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Ruaniaceae;
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
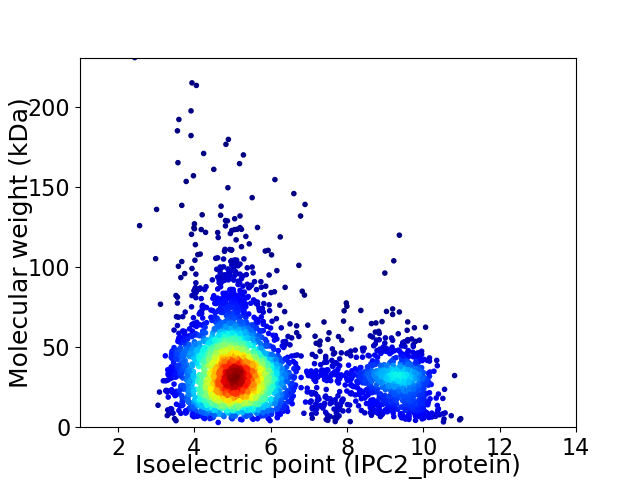
Virtual 2D-PAGE plot for 4192 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H5FA42|A0A1H5FA42_9MICO PucR C-terminal helix-turn-helix domain-containing protein OS=Haloactinobacterium album OX=648782 GN=SAMN04488554_1279 PE=3 SV=1
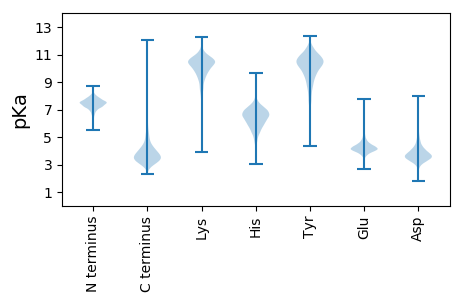
MM1 pKa = 7.38SRR3 pKa = 11.84RR4 pKa = 11.84ARR6 pKa = 11.84WRR8 pKa = 11.84RR9 pKa = 11.84AAGVFTVALLVATGLAAVEE28 pKa = 4.23ATVAPAPAASAASSPDD44 pKa = 2.94GTGIRR49 pKa = 11.84QSQTFYY55 pKa = 10.97AYY57 pKa = 10.28ADD59 pKa = 3.5AGEE62 pKa = 4.37TVSAAFEE69 pKa = 4.44VVNRR73 pKa = 11.84WSFTGSAVFTVTAPDD88 pKa = 4.34GATQTCTTPAGAALGSVCEE107 pKa = 4.22IGPMTSEE114 pKa = 4.1TSGVWTVDD122 pKa = 2.37VHH124 pKa = 7.59RR125 pKa = 11.84PGDD128 pKa = 3.5FGGAFTWDD136 pKa = 2.92ITVADD141 pKa = 4.94GGTAIPGRR149 pKa = 11.84VWTDD153 pKa = 2.88RR154 pKa = 11.84FLMWNRR160 pKa = 11.84EE161 pKa = 4.06FVDD164 pKa = 3.53LTLWYY169 pKa = 10.17VGASGHH175 pKa = 5.34QYY177 pKa = 9.04EE178 pKa = 4.45VSYY181 pKa = 11.17FDD183 pKa = 3.96FYY185 pKa = 11.39GIEE188 pKa = 4.3SLFSSNQFGVVYY200 pKa = 10.7DD201 pKa = 4.24GTCEE205 pKa = 3.76SAYY208 pKa = 10.52QSAEE212 pKa = 3.88DD213 pKa = 4.36RR214 pKa = 11.84PSGPYY219 pKa = 10.13YY220 pKa = 10.15DD221 pKa = 5.38DD222 pKa = 4.44HH223 pKa = 8.22LSNSGQEE230 pKa = 4.33CGPLYY235 pKa = 11.36NMFFAPPAADD245 pKa = 4.22LPASAADD252 pKa = 3.52ATGATQFILPPVQPPTVSEE271 pKa = 4.12LRR273 pKa = 11.84FTPDD277 pKa = 2.79TPDD280 pKa = 3.09TRR282 pKa = 11.84EE283 pKa = 3.94GTFSFEE289 pKa = 4.2TNHH292 pKa = 6.12VGNADD297 pKa = 4.1LLIDD301 pKa = 4.02ADD303 pKa = 4.5GNGSYY308 pKa = 11.03DD309 pKa = 3.55DD310 pKa = 4.02TVDD313 pKa = 2.98RR314 pKa = 11.84TIPVSVNTGAEE325 pKa = 3.82PSVAFDD331 pKa = 3.79GLDD334 pKa = 3.59GEE336 pKa = 5.33GNAISPSTRR345 pKa = 11.84FNATMRR351 pKa = 11.84ITQAGEE357 pKa = 3.45IHH359 pKa = 6.4FVNGDD364 pKa = 3.3VEE366 pKa = 4.65SRR368 pKa = 11.84GGISVTALTGTADD381 pKa = 3.88GTSTLYY387 pKa = 10.57WNDD390 pKa = 3.53SEE392 pKa = 4.99LQTEE396 pKa = 4.54EE397 pKa = 5.13RR398 pKa = 11.84DD399 pKa = 3.91QSCLPPVLDD408 pKa = 4.73GRR410 pKa = 11.84DD411 pKa = 3.9GQDD414 pKa = 2.83SSVSGGVHH422 pKa = 6.54GWPCTPRR429 pKa = 11.84NPNDD433 pKa = 3.56GVTGPWGDD441 pKa = 3.43VRR443 pKa = 11.84LIDD446 pKa = 4.27DD447 pKa = 3.61WTFNPVDD454 pKa = 3.52VSASVEE460 pKa = 3.87IPAAEE465 pKa = 4.06GNYY468 pKa = 9.68VVTKK472 pKa = 10.75SSDD475 pKa = 3.82PEE477 pKa = 4.68PGTAVHH483 pKa = 6.77PGDD486 pKa = 3.46VVTYY490 pKa = 8.45TIEE493 pKa = 3.9VAQIGNDD500 pKa = 3.19SAAANVSDD508 pKa = 5.1DD509 pKa = 4.41LSDD512 pKa = 3.9VLDD515 pKa = 4.32DD516 pKa = 4.41ASYY519 pKa = 11.51NGDD522 pKa = 3.28ASATAGEE529 pKa = 4.23VDD531 pKa = 3.74VTGTTLNWNGTLLDD545 pKa = 4.29GGSARR550 pKa = 11.84ITYY553 pKa = 10.29SVTVDD558 pKa = 3.45DD559 pKa = 5.57AGDD562 pKa = 3.46VSSGDD567 pKa = 3.56GQVGNVVTSPGCGSDD582 pKa = 3.21ACATEE587 pKa = 4.73HH588 pKa = 6.74PVGDD592 pKa = 3.74YY593 pKa = 10.8SVEE596 pKa = 4.04KK597 pKa = 10.39SAEE600 pKa = 4.03PAPGSAVTDD609 pKa = 4.03GDD611 pKa = 4.09TVAYY615 pKa = 8.03TLTVAQEE622 pKa = 3.68GAAAVEE628 pKa = 4.62GASLVDD634 pKa = 4.34DD635 pKa = 5.02LSDD638 pKa = 3.78VLDD641 pKa = 4.24DD642 pKa = 4.68AVWNGDD648 pKa = 3.48LVASAGAAEE657 pKa = 4.25FDD659 pKa = 3.85EE660 pKa = 5.23DD661 pKa = 3.85AQEE664 pKa = 4.31LRR666 pKa = 11.84WSGDD670 pKa = 3.18LGIDD674 pKa = 3.51DD675 pKa = 4.95VVTISYY681 pKa = 10.85SVMVTGSGDD690 pKa = 3.38TSVDD694 pKa = 3.12NTVASDD700 pKa = 3.84GCGTAEE706 pKa = 4.31EE707 pKa = 4.92CQSSHH712 pKa = 7.19PYY714 pKa = 9.86GGYY717 pKa = 10.55GVVKK721 pKa = 10.36SADD724 pKa = 3.64PAPGGGVAEE733 pKa = 4.82GEE735 pKa = 4.55TVAYY739 pKa = 8.26TLTVTQSGQGSVPAEE754 pKa = 4.02LSDD757 pKa = 5.13DD758 pKa = 3.73LTAVLDD764 pKa = 4.0DD765 pKa = 4.49AVYY768 pKa = 11.19NDD770 pKa = 5.08DD771 pKa = 4.17LQASAGAASYY781 pKa = 10.79DD782 pKa = 4.13SEE784 pKa = 5.13SGTIDD789 pKa = 2.87WDD791 pKa = 3.63GTLARR796 pKa = 11.84GDD798 pKa = 3.88VVTISYY804 pKa = 10.54SVTVQPVGTGDD815 pKa = 3.16GSLRR819 pKa = 11.84NVVTSPGCDD828 pKa = 2.96SDD830 pKa = 4.36ADD832 pKa = 3.92CVTEE836 pKa = 3.79NRR838 pKa = 11.84VGGFVYY844 pKa = 10.52SKK846 pKa = 10.69SADD849 pKa = 3.58GGPGSQVSQGDD860 pKa = 3.98VITYY864 pKa = 10.08SVTVTHH870 pKa = 6.93TGAGPVAEE878 pKa = 4.76ARR880 pKa = 11.84VSDD883 pKa = 4.33DD884 pKa = 3.64LSGVLDD890 pKa = 4.4DD891 pKa = 5.26ATFNDD896 pKa = 4.46DD897 pKa = 3.65ASASSGSIRR906 pKa = 11.84VDD908 pKa = 3.0GSTLDD913 pKa = 3.49WTGSLAPGDD922 pKa = 3.96VVEE925 pKa = 4.51ITYY928 pKa = 10.86SVTVTGAGDD937 pKa = 3.6LAVDD941 pKa = 3.83NVVLTDD947 pKa = 4.15DD948 pKa = 4.16PRR950 pKa = 11.84GVCDD954 pKa = 4.75PDD956 pKa = 3.45ATCATEE962 pKa = 4.26HH963 pKa = 5.74QVPPSPDD970 pKa = 3.24LPATGARR977 pKa = 11.84VGAWVIPAALLMTGLGALVLMGARR1001 pKa = 11.84RR1002 pKa = 11.84HH1003 pKa = 5.5RR1004 pKa = 11.84AAEE1007 pKa = 4.02KK1008 pKa = 10.58
MM1 pKa = 7.38SRR3 pKa = 11.84RR4 pKa = 11.84ARR6 pKa = 11.84WRR8 pKa = 11.84RR9 pKa = 11.84AAGVFTVALLVATGLAAVEE28 pKa = 4.23ATVAPAPAASAASSPDD44 pKa = 2.94GTGIRR49 pKa = 11.84QSQTFYY55 pKa = 10.97AYY57 pKa = 10.28ADD59 pKa = 3.5AGEE62 pKa = 4.37TVSAAFEE69 pKa = 4.44VVNRR73 pKa = 11.84WSFTGSAVFTVTAPDD88 pKa = 4.34GATQTCTTPAGAALGSVCEE107 pKa = 4.22IGPMTSEE114 pKa = 4.1TSGVWTVDD122 pKa = 2.37VHH124 pKa = 7.59RR125 pKa = 11.84PGDD128 pKa = 3.5FGGAFTWDD136 pKa = 2.92ITVADD141 pKa = 4.94GGTAIPGRR149 pKa = 11.84VWTDD153 pKa = 2.88RR154 pKa = 11.84FLMWNRR160 pKa = 11.84EE161 pKa = 4.06FVDD164 pKa = 3.53LTLWYY169 pKa = 10.17VGASGHH175 pKa = 5.34QYY177 pKa = 9.04EE178 pKa = 4.45VSYY181 pKa = 11.17FDD183 pKa = 3.96FYY185 pKa = 11.39GIEE188 pKa = 4.3SLFSSNQFGVVYY200 pKa = 10.7DD201 pKa = 4.24GTCEE205 pKa = 3.76SAYY208 pKa = 10.52QSAEE212 pKa = 3.88DD213 pKa = 4.36RR214 pKa = 11.84PSGPYY219 pKa = 10.13YY220 pKa = 10.15DD221 pKa = 5.38DD222 pKa = 4.44HH223 pKa = 8.22LSNSGQEE230 pKa = 4.33CGPLYY235 pKa = 11.36NMFFAPPAADD245 pKa = 4.22LPASAADD252 pKa = 3.52ATGATQFILPPVQPPTVSEE271 pKa = 4.12LRR273 pKa = 11.84FTPDD277 pKa = 2.79TPDD280 pKa = 3.09TRR282 pKa = 11.84EE283 pKa = 3.94GTFSFEE289 pKa = 4.2TNHH292 pKa = 6.12VGNADD297 pKa = 4.1LLIDD301 pKa = 4.02ADD303 pKa = 4.5GNGSYY308 pKa = 11.03DD309 pKa = 3.55DD310 pKa = 4.02TVDD313 pKa = 2.98RR314 pKa = 11.84TIPVSVNTGAEE325 pKa = 3.82PSVAFDD331 pKa = 3.79GLDD334 pKa = 3.59GEE336 pKa = 5.33GNAISPSTRR345 pKa = 11.84FNATMRR351 pKa = 11.84ITQAGEE357 pKa = 3.45IHH359 pKa = 6.4FVNGDD364 pKa = 3.3VEE366 pKa = 4.65SRR368 pKa = 11.84GGISVTALTGTADD381 pKa = 3.88GTSTLYY387 pKa = 10.57WNDD390 pKa = 3.53SEE392 pKa = 4.99LQTEE396 pKa = 4.54EE397 pKa = 5.13RR398 pKa = 11.84DD399 pKa = 3.91QSCLPPVLDD408 pKa = 4.73GRR410 pKa = 11.84DD411 pKa = 3.9GQDD414 pKa = 2.83SSVSGGVHH422 pKa = 6.54GWPCTPRR429 pKa = 11.84NPNDD433 pKa = 3.56GVTGPWGDD441 pKa = 3.43VRR443 pKa = 11.84LIDD446 pKa = 4.27DD447 pKa = 3.61WTFNPVDD454 pKa = 3.52VSASVEE460 pKa = 3.87IPAAEE465 pKa = 4.06GNYY468 pKa = 9.68VVTKK472 pKa = 10.75SSDD475 pKa = 3.82PEE477 pKa = 4.68PGTAVHH483 pKa = 6.77PGDD486 pKa = 3.46VVTYY490 pKa = 8.45TIEE493 pKa = 3.9VAQIGNDD500 pKa = 3.19SAAANVSDD508 pKa = 5.1DD509 pKa = 4.41LSDD512 pKa = 3.9VLDD515 pKa = 4.32DD516 pKa = 4.41ASYY519 pKa = 11.51NGDD522 pKa = 3.28ASATAGEE529 pKa = 4.23VDD531 pKa = 3.74VTGTTLNWNGTLLDD545 pKa = 4.29GGSARR550 pKa = 11.84ITYY553 pKa = 10.29SVTVDD558 pKa = 3.45DD559 pKa = 5.57AGDD562 pKa = 3.46VSSGDD567 pKa = 3.56GQVGNVVTSPGCGSDD582 pKa = 3.21ACATEE587 pKa = 4.73HH588 pKa = 6.74PVGDD592 pKa = 3.74YY593 pKa = 10.8SVEE596 pKa = 4.04KK597 pKa = 10.39SAEE600 pKa = 4.03PAPGSAVTDD609 pKa = 4.03GDD611 pKa = 4.09TVAYY615 pKa = 8.03TLTVAQEE622 pKa = 3.68GAAAVEE628 pKa = 4.62GASLVDD634 pKa = 4.34DD635 pKa = 5.02LSDD638 pKa = 3.78VLDD641 pKa = 4.24DD642 pKa = 4.68AVWNGDD648 pKa = 3.48LVASAGAAEE657 pKa = 4.25FDD659 pKa = 3.85EE660 pKa = 5.23DD661 pKa = 3.85AQEE664 pKa = 4.31LRR666 pKa = 11.84WSGDD670 pKa = 3.18LGIDD674 pKa = 3.51DD675 pKa = 4.95VVTISYY681 pKa = 10.85SVMVTGSGDD690 pKa = 3.38TSVDD694 pKa = 3.12NTVASDD700 pKa = 3.84GCGTAEE706 pKa = 4.31EE707 pKa = 4.92CQSSHH712 pKa = 7.19PYY714 pKa = 9.86GGYY717 pKa = 10.55GVVKK721 pKa = 10.36SADD724 pKa = 3.64PAPGGGVAEE733 pKa = 4.82GEE735 pKa = 4.55TVAYY739 pKa = 8.26TLTVTQSGQGSVPAEE754 pKa = 4.02LSDD757 pKa = 5.13DD758 pKa = 3.73LTAVLDD764 pKa = 4.0DD765 pKa = 4.49AVYY768 pKa = 11.19NDD770 pKa = 5.08DD771 pKa = 4.17LQASAGAASYY781 pKa = 10.79DD782 pKa = 4.13SEE784 pKa = 5.13SGTIDD789 pKa = 2.87WDD791 pKa = 3.63GTLARR796 pKa = 11.84GDD798 pKa = 3.88VVTISYY804 pKa = 10.54SVTVQPVGTGDD815 pKa = 3.16GSLRR819 pKa = 11.84NVVTSPGCDD828 pKa = 2.96SDD830 pKa = 4.36ADD832 pKa = 3.92CVTEE836 pKa = 3.79NRR838 pKa = 11.84VGGFVYY844 pKa = 10.52SKK846 pKa = 10.69SADD849 pKa = 3.58GGPGSQVSQGDD860 pKa = 3.98VITYY864 pKa = 10.08SVTVTHH870 pKa = 6.93TGAGPVAEE878 pKa = 4.76ARR880 pKa = 11.84VSDD883 pKa = 4.33DD884 pKa = 3.64LSGVLDD890 pKa = 4.4DD891 pKa = 5.26ATFNDD896 pKa = 4.46DD897 pKa = 3.65ASASSGSIRR906 pKa = 11.84VDD908 pKa = 3.0GSTLDD913 pKa = 3.49WTGSLAPGDD922 pKa = 3.96VVEE925 pKa = 4.51ITYY928 pKa = 10.86SVTVTGAGDD937 pKa = 3.6LAVDD941 pKa = 3.83NVVLTDD947 pKa = 4.15DD948 pKa = 4.16PRR950 pKa = 11.84GVCDD954 pKa = 4.75PDD956 pKa = 3.45ATCATEE962 pKa = 4.26HH963 pKa = 5.74QVPPSPDD970 pKa = 3.24LPATGARR977 pKa = 11.84VGAWVIPAALLMTGLGALVLMGARR1001 pKa = 11.84RR1002 pKa = 11.84HH1003 pKa = 5.5RR1004 pKa = 11.84AAEE1007 pKa = 4.02KK1008 pKa = 10.58
Molecular weight: 103.54 kDa
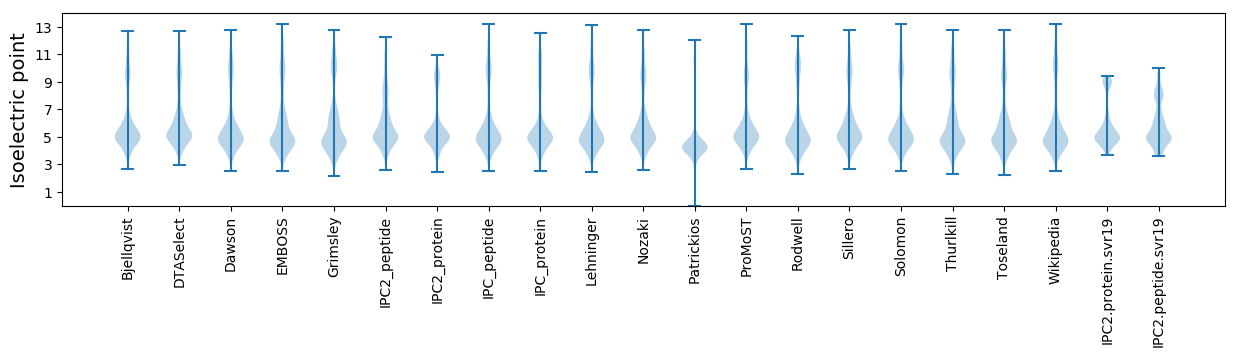
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H5LNV2|A0A1H5LNV2_9MICO TIR domain-containing protein OS=Haloactinobacterium album OX=648782 GN=SAMN04488554_2902 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.97GRR40 pKa = 11.84TEE42 pKa = 3.99LSAA45 pKa = 4.86
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.97GRR40 pKa = 11.84TEE42 pKa = 3.99LSAA45 pKa = 4.86
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1448774 |
29 |
2441 |
345.6 |
37.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.017 ± 0.05 | 0.611 ± 0.009 |
6.407 ± 0.043 | 5.96 ± 0.036 |
2.844 ± 0.022 | 9.119 ± 0.042 |
2.261 ± 0.021 | 3.902 ± 0.026 |
1.314 ± 0.022 | 10.074 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.895 ± 0.016 | 1.876 ± 0.019 |
5.764 ± 0.03 | 2.994 ± 0.017 |
7.247 ± 0.052 | 5.488 ± 0.027 |
6.569 ± 0.037 | 8.957 ± 0.038 |
1.645 ± 0.019 | 2.057 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
