
Solimonas fluminis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Nevskiales; Sinobacteraceae; Solimonas
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
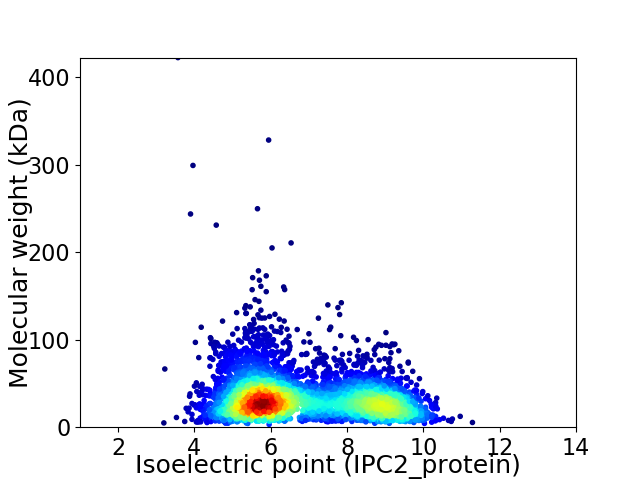
Virtual 2D-PAGE plot for 4276 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S5TK49|A0A2S5TK49_9GAMM Urease subunit gamma OS=Solimonas fluminis OX=2086571 GN=ureA PE=3 SV=1
MM1 pKa = 7.12NRR3 pKa = 11.84VGKK6 pKa = 8.21ATRR9 pKa = 11.84WASALMAVGAAGAMPVAQADD29 pKa = 3.73IEE31 pKa = 4.47LGNGLSVTGFLDD43 pKa = 3.34MSYY46 pKa = 11.26SSVNPDD52 pKa = 2.98SGSSTEE58 pKa = 4.6SVGIDD63 pKa = 3.41QFEE66 pKa = 4.24MDD68 pKa = 4.69FKK70 pKa = 10.74YY71 pKa = 10.52AGSGGVSAQVDD82 pKa = 3.42IEE84 pKa = 4.22YY85 pKa = 11.24GEE87 pKa = 4.76GFDD90 pKa = 5.35GSDD93 pKa = 3.42DD94 pKa = 3.68EE95 pKa = 5.05TFVEE99 pKa = 4.49QAFITKK105 pKa = 10.32AFTEE109 pKa = 4.26KK110 pKa = 10.64FSVKK114 pKa = 10.11AGRR117 pKa = 11.84FLSYY121 pKa = 10.62SGWEE125 pKa = 4.17TEE127 pKa = 4.12EE128 pKa = 3.92PTGLFQYY135 pKa = 10.72SGTGYY140 pKa = 10.75AKK142 pKa = 10.45YY143 pKa = 10.07FYY145 pKa = 10.46GYY147 pKa = 8.58YY148 pKa = 9.85QNGVSAYY155 pKa = 9.1YY156 pKa = 10.43NGGKK160 pKa = 9.13FAVMGSAVTSAFNPNDD176 pKa = 3.64RR177 pKa = 11.84NNVEE181 pKa = 4.24GVDD184 pKa = 3.74DD185 pKa = 4.08KK186 pKa = 11.5KK187 pKa = 11.32GYY189 pKa = 10.25EE190 pKa = 3.97FGLAVMPIEE199 pKa = 4.54GLTAKK204 pKa = 10.48AFYY207 pKa = 10.36IADD210 pKa = 3.91NDD212 pKa = 3.77TDD214 pKa = 3.67TDD216 pKa = 4.21IINVWVSYY224 pKa = 10.69AVAGFTFAGEE234 pKa = 4.4YY235 pKa = 8.67NTVEE239 pKa = 4.33YY240 pKa = 10.23GDD242 pKa = 3.92SSAVVDD248 pKa = 4.3GDD250 pKa = 3.64GDD252 pKa = 4.27GYY254 pKa = 11.79LLMANYY260 pKa = 10.73AKK262 pKa = 10.43GPYY265 pKa = 9.91GITLRR270 pKa = 11.84YY271 pKa = 9.32HH272 pKa = 6.92DD273 pKa = 6.08FEE275 pKa = 5.36IQDD278 pKa = 3.55AGGATVDD285 pKa = 4.31DD286 pKa = 5.07GSAITLSPSYY296 pKa = 10.56KK297 pKa = 10.35VADD300 pKa = 3.62NLLLVGEE307 pKa = 4.26YY308 pKa = 10.76RR309 pKa = 11.84MDD311 pKa = 3.68EE312 pKa = 4.12SDD314 pKa = 3.65TNGDD318 pKa = 3.38SDD320 pKa = 4.01SFAIEE325 pKa = 3.79ALFTFF330 pKa = 5.15
MM1 pKa = 7.12NRR3 pKa = 11.84VGKK6 pKa = 8.21ATRR9 pKa = 11.84WASALMAVGAAGAMPVAQADD29 pKa = 3.73IEE31 pKa = 4.47LGNGLSVTGFLDD43 pKa = 3.34MSYY46 pKa = 11.26SSVNPDD52 pKa = 2.98SGSSTEE58 pKa = 4.6SVGIDD63 pKa = 3.41QFEE66 pKa = 4.24MDD68 pKa = 4.69FKK70 pKa = 10.74YY71 pKa = 10.52AGSGGVSAQVDD82 pKa = 3.42IEE84 pKa = 4.22YY85 pKa = 11.24GEE87 pKa = 4.76GFDD90 pKa = 5.35GSDD93 pKa = 3.42DD94 pKa = 3.68EE95 pKa = 5.05TFVEE99 pKa = 4.49QAFITKK105 pKa = 10.32AFTEE109 pKa = 4.26KK110 pKa = 10.64FSVKK114 pKa = 10.11AGRR117 pKa = 11.84FLSYY121 pKa = 10.62SGWEE125 pKa = 4.17TEE127 pKa = 4.12EE128 pKa = 3.92PTGLFQYY135 pKa = 10.72SGTGYY140 pKa = 10.75AKK142 pKa = 10.45YY143 pKa = 10.07FYY145 pKa = 10.46GYY147 pKa = 8.58YY148 pKa = 9.85QNGVSAYY155 pKa = 9.1YY156 pKa = 10.43NGGKK160 pKa = 9.13FAVMGSAVTSAFNPNDD176 pKa = 3.64RR177 pKa = 11.84NNVEE181 pKa = 4.24GVDD184 pKa = 3.74DD185 pKa = 4.08KK186 pKa = 11.5KK187 pKa = 11.32GYY189 pKa = 10.25EE190 pKa = 3.97FGLAVMPIEE199 pKa = 4.54GLTAKK204 pKa = 10.48AFYY207 pKa = 10.36IADD210 pKa = 3.91NDD212 pKa = 3.77TDD214 pKa = 3.67TDD216 pKa = 4.21IINVWVSYY224 pKa = 10.69AVAGFTFAGEE234 pKa = 4.4YY235 pKa = 8.67NTVEE239 pKa = 4.33YY240 pKa = 10.23GDD242 pKa = 3.92SSAVVDD248 pKa = 4.3GDD250 pKa = 3.64GDD252 pKa = 4.27GYY254 pKa = 11.79LLMANYY260 pKa = 10.73AKK262 pKa = 10.43GPYY265 pKa = 9.91GITLRR270 pKa = 11.84YY271 pKa = 9.32HH272 pKa = 6.92DD273 pKa = 6.08FEE275 pKa = 5.36IQDD278 pKa = 3.55AGGATVDD285 pKa = 4.31DD286 pKa = 5.07GSAITLSPSYY296 pKa = 10.56KK297 pKa = 10.35VADD300 pKa = 3.62NLLLVGEE307 pKa = 4.26YY308 pKa = 10.76RR309 pKa = 11.84MDD311 pKa = 3.68EE312 pKa = 4.12SDD314 pKa = 3.65TNGDD318 pKa = 3.38SDD320 pKa = 4.01SFAIEE325 pKa = 3.79ALFTFF330 pKa = 5.15
Molecular weight: 35.36 kDa
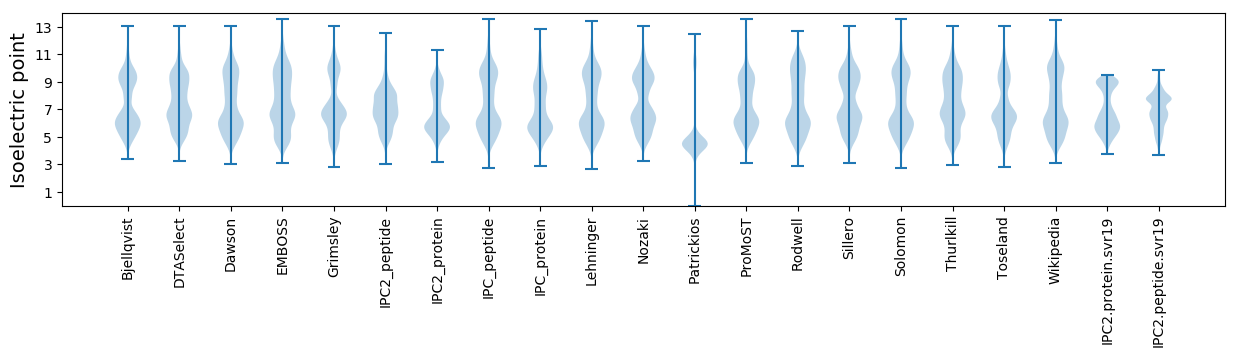
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S5TJZ1|A0A2S5TJZ1_9GAMM Enoyl-CoA hydratase OS=Solimonas fluminis OX=2086571 GN=C3942_06335 PE=3 SV=1
MM1 pKa = 6.59ATKK4 pKa = 9.57RR5 pKa = 11.84TFQPKK10 pKa = 8.81KK11 pKa = 8.99LRR13 pKa = 11.84RR14 pKa = 11.84ARR16 pKa = 11.84THH18 pKa = 5.92GFRR21 pKa = 11.84ARR23 pKa = 11.84MATVGGRR30 pKa = 11.84KK31 pKa = 9.31VISRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 9.94GRR41 pKa = 11.84KK42 pKa = 8.89RR43 pKa = 11.84LIPP46 pKa = 4.02
MM1 pKa = 6.59ATKK4 pKa = 9.57RR5 pKa = 11.84TFQPKK10 pKa = 8.81KK11 pKa = 8.99LRR13 pKa = 11.84RR14 pKa = 11.84ARR16 pKa = 11.84THH18 pKa = 5.92GFRR21 pKa = 11.84ARR23 pKa = 11.84MATVGGRR30 pKa = 11.84KK31 pKa = 9.31VISRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 9.94GRR41 pKa = 11.84KK42 pKa = 8.89RR43 pKa = 11.84LIPP46 pKa = 4.02
Molecular weight: 5.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1399617 |
29 |
4449 |
327.3 |
35.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.38 ± 0.054 | 0.953 ± 0.013 |
5.316 ± 0.029 | 5.809 ± 0.036 |
3.444 ± 0.022 | 8.81 ± 0.047 |
2.092 ± 0.021 | 4.189 ± 0.028 |
3.128 ± 0.034 | 11.069 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.242 ± 0.021 | 2.531 ± 0.026 |
5.578 ± 0.036 | 3.926 ± 0.022 |
7.666 ± 0.038 | 5.267 ± 0.034 |
4.629 ± 0.039 | 7.006 ± 0.031 |
1.483 ± 0.017 | 2.481 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
