
Avian paraavulavirus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Paraavulavirus
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
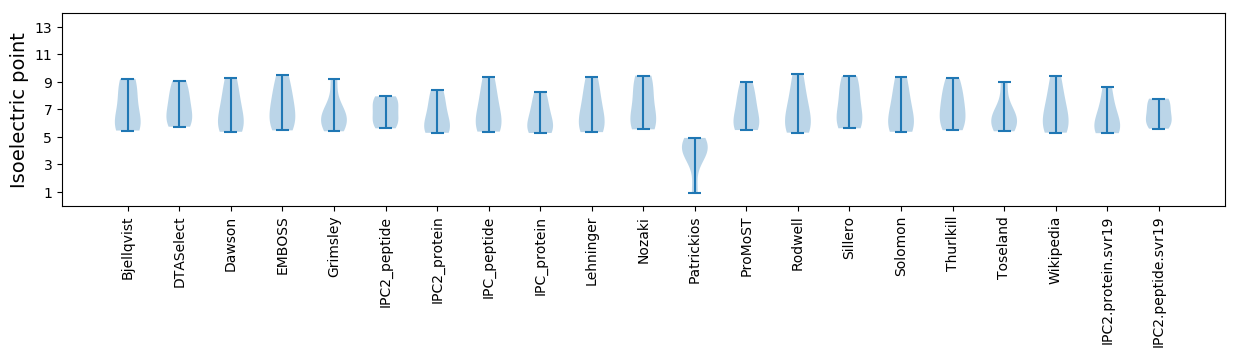
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|D5FGX3|D5FGX3_9MONO Phosphoprotein OS=Avian paraavulavirus 3 OX=2560327 PE=4 SV=1
MM1 pKa = 7.51AGIFNTYY8 pKa = 8.9EE9 pKa = 4.47LFVKK13 pKa = 10.42DD14 pKa = 3.59QTCMHH19 pKa = 6.52KK20 pKa = 10.31RR21 pKa = 11.84AASLISGGQLKK32 pKa = 11.04SNIPVFITTRR42 pKa = 11.84DD43 pKa = 3.64DD44 pKa = 3.86PAVRR48 pKa = 11.84WNLVCFNLRR57 pKa = 11.84LIVSEE62 pKa = 4.42SSTSVIRR69 pKa = 11.84QGAMISLLSVTASNMRR85 pKa = 11.84ALAAIAGQTDD95 pKa = 3.12EE96 pKa = 4.87SMINIIEE103 pKa = 4.22VVDD106 pKa = 4.23FNGLEE111 pKa = 4.17PQCDD115 pKa = 3.61PRR117 pKa = 11.84SGLDD121 pKa = 3.31AQKK124 pKa = 10.97QDD126 pKa = 3.37MFKK129 pKa = 10.79DD130 pKa = 3.46IASDD134 pKa = 3.56MPKK137 pKa = 10.18VLGSGTPFQNVSAEE151 pKa = 4.35TNNPEE156 pKa = 4.0DD157 pKa = 2.71THH159 pKa = 5.91MFLRR163 pKa = 11.84SAISVLTQIWILVAKK178 pKa = 10.57AMTNIEE184 pKa = 4.21GSHH187 pKa = 5.58EE188 pKa = 4.02ASDD191 pKa = 3.71RR192 pKa = 11.84RR193 pKa = 11.84LAKK196 pKa = 9.15YY197 pKa = 6.8TQQNRR202 pKa = 11.84IDD204 pKa = 3.84RR205 pKa = 11.84RR206 pKa = 11.84FMLAQATRR214 pKa = 11.84TACQQIIKK222 pKa = 10.37DD223 pKa = 3.65SLTIRR228 pKa = 11.84RR229 pKa = 11.84FLVTEE234 pKa = 4.05LRR236 pKa = 11.84KK237 pKa = 10.22SRR239 pKa = 11.84GALHH243 pKa = 6.61SGSSYY248 pKa = 10.2YY249 pKa = 11.61AMVGDD254 pKa = 3.76MQAYY258 pKa = 9.01IFNAGLTPFLTTLRR272 pKa = 11.84YY273 pKa = 10.01GIGTKK278 pKa = 9.71YY279 pKa = 9.83HH280 pKa = 6.69ALAISSLTGDD290 pKa = 3.69LNKK293 pKa = 10.25IKK295 pKa = 10.77GLLTLYY301 pKa = 10.44KK302 pKa = 10.41EE303 pKa = 4.44KK304 pKa = 10.97GSDD307 pKa = 3.31AGYY310 pKa = 8.59MALLEE315 pKa = 5.34DD316 pKa = 4.72ADD318 pKa = 4.33CMQFAPGNYY327 pKa = 9.65ALLYY331 pKa = 9.13SYY333 pKa = 11.68AMGVASVHH341 pKa = 6.57DD342 pKa = 4.01EE343 pKa = 3.81GMRR346 pKa = 11.84NYY348 pKa = 10.34QYY350 pKa = 11.66ARR352 pKa = 11.84RR353 pKa = 11.84FLHH356 pKa = 6.34KK357 pKa = 10.55GMYY360 pKa = 9.57QFGRR364 pKa = 11.84DD365 pKa = 2.85IATQHH370 pKa = 4.81QHH372 pKa = 6.96ALDD375 pKa = 3.76EE376 pKa = 4.51SLAQEE381 pKa = 4.08MRR383 pKa = 11.84ITEE386 pKa = 4.09ADD388 pKa = 3.74RR389 pKa = 11.84ANLKK393 pKa = 10.76VMMANIGEE401 pKa = 4.21ASHH404 pKa = 6.71YY405 pKa = 10.95SDD407 pKa = 4.53IPSAGPSGIPAFNDD421 pKa = 3.37PPEE424 pKa = 4.14EE425 pKa = 4.0LFGEE429 pKa = 4.25PSYY432 pKa = 11.23RR433 pKa = 11.84KK434 pKa = 9.91LPEE437 pKa = 4.05EE438 pKa = 4.08PQVVEE443 pKa = 4.38LQDD446 pKa = 4.46RR447 pKa = 11.84DD448 pKa = 4.56DD449 pKa = 5.18DD450 pKa = 4.2EE451 pKa = 4.75QDD453 pKa = 3.42EE454 pKa = 4.6YY455 pKa = 12.1DD456 pKa = 3.37MM457 pKa = 6.54
MM1 pKa = 7.51AGIFNTYY8 pKa = 8.9EE9 pKa = 4.47LFVKK13 pKa = 10.42DD14 pKa = 3.59QTCMHH19 pKa = 6.52KK20 pKa = 10.31RR21 pKa = 11.84AASLISGGQLKK32 pKa = 11.04SNIPVFITTRR42 pKa = 11.84DD43 pKa = 3.64DD44 pKa = 3.86PAVRR48 pKa = 11.84WNLVCFNLRR57 pKa = 11.84LIVSEE62 pKa = 4.42SSTSVIRR69 pKa = 11.84QGAMISLLSVTASNMRR85 pKa = 11.84ALAAIAGQTDD95 pKa = 3.12EE96 pKa = 4.87SMINIIEE103 pKa = 4.22VVDD106 pKa = 4.23FNGLEE111 pKa = 4.17PQCDD115 pKa = 3.61PRR117 pKa = 11.84SGLDD121 pKa = 3.31AQKK124 pKa = 10.97QDD126 pKa = 3.37MFKK129 pKa = 10.79DD130 pKa = 3.46IASDD134 pKa = 3.56MPKK137 pKa = 10.18VLGSGTPFQNVSAEE151 pKa = 4.35TNNPEE156 pKa = 4.0DD157 pKa = 2.71THH159 pKa = 5.91MFLRR163 pKa = 11.84SAISVLTQIWILVAKK178 pKa = 10.57AMTNIEE184 pKa = 4.21GSHH187 pKa = 5.58EE188 pKa = 4.02ASDD191 pKa = 3.71RR192 pKa = 11.84RR193 pKa = 11.84LAKK196 pKa = 9.15YY197 pKa = 6.8TQQNRR202 pKa = 11.84IDD204 pKa = 3.84RR205 pKa = 11.84RR206 pKa = 11.84FMLAQATRR214 pKa = 11.84TACQQIIKK222 pKa = 10.37DD223 pKa = 3.65SLTIRR228 pKa = 11.84RR229 pKa = 11.84FLVTEE234 pKa = 4.05LRR236 pKa = 11.84KK237 pKa = 10.22SRR239 pKa = 11.84GALHH243 pKa = 6.61SGSSYY248 pKa = 10.2YY249 pKa = 11.61AMVGDD254 pKa = 3.76MQAYY258 pKa = 9.01IFNAGLTPFLTTLRR272 pKa = 11.84YY273 pKa = 10.01GIGTKK278 pKa = 9.71YY279 pKa = 9.83HH280 pKa = 6.69ALAISSLTGDD290 pKa = 3.69LNKK293 pKa = 10.25IKK295 pKa = 10.77GLLTLYY301 pKa = 10.44KK302 pKa = 10.41EE303 pKa = 4.44KK304 pKa = 10.97GSDD307 pKa = 3.31AGYY310 pKa = 8.59MALLEE315 pKa = 5.34DD316 pKa = 4.72ADD318 pKa = 4.33CMQFAPGNYY327 pKa = 9.65ALLYY331 pKa = 9.13SYY333 pKa = 11.68AMGVASVHH341 pKa = 6.57DD342 pKa = 4.01EE343 pKa = 3.81GMRR346 pKa = 11.84NYY348 pKa = 10.34QYY350 pKa = 11.66ARR352 pKa = 11.84RR353 pKa = 11.84FLHH356 pKa = 6.34KK357 pKa = 10.55GMYY360 pKa = 9.57QFGRR364 pKa = 11.84DD365 pKa = 2.85IATQHH370 pKa = 4.81QHH372 pKa = 6.96ALDD375 pKa = 3.76EE376 pKa = 4.51SLAQEE381 pKa = 4.08MRR383 pKa = 11.84ITEE386 pKa = 4.09ADD388 pKa = 3.74RR389 pKa = 11.84ANLKK393 pKa = 10.76VMMANIGEE401 pKa = 4.21ASHH404 pKa = 6.71YY405 pKa = 10.95SDD407 pKa = 4.53IPSAGPSGIPAFNDD421 pKa = 3.37PPEE424 pKa = 4.14EE425 pKa = 4.0LFGEE429 pKa = 4.25PSYY432 pKa = 11.23RR433 pKa = 11.84KK434 pKa = 9.91LPEE437 pKa = 4.05EE438 pKa = 4.08PQVVEE443 pKa = 4.38LQDD446 pKa = 4.46RR447 pKa = 11.84DD448 pKa = 4.56DD449 pKa = 5.18DD450 pKa = 4.2EE451 pKa = 4.75QDD453 pKa = 3.42EE454 pKa = 4.6YY455 pKa = 12.1DD456 pKa = 3.37MM457 pKa = 6.54
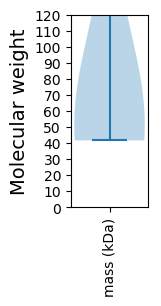
Molecular weight: 50.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D5FGX5|D5FGX5_9MONO Fusion glycoprotein F0 OS=Avian paraavulavirus 3 OX=2560327 GN=F PE=3 SV=1
MM1 pKa = 7.55AAHH4 pKa = 7.22PNHH7 pKa = 6.99ANPSSSISLMHH18 pKa = 7.63DD19 pKa = 3.5DD20 pKa = 5.63PSIQTQLLAFPLISEE35 pKa = 4.31KK36 pKa = 10.54TEE38 pKa = 3.77TGTTKK43 pKa = 10.57LQPQVRR49 pKa = 11.84MQSFLSTDD57 pKa = 3.07SQKK60 pKa = 10.89YY61 pKa = 9.26HH62 pKa = 6.38LVFINTYY69 pKa = 10.34GFIAEE74 pKa = 5.21DD75 pKa = 4.69FNCSPTNGFVPALFQPKK92 pKa = 10.21SKK94 pKa = 10.29VLSSAMVTLGAVPADD109 pKa = 3.67TVLQDD114 pKa = 3.67LQKK117 pKa = 11.21DD118 pKa = 4.44LIAMRR123 pKa = 11.84FKK125 pKa = 10.78VRR127 pKa = 11.84KK128 pKa = 8.73SASAKK133 pKa = 10.0EE134 pKa = 4.36LILFSTDD141 pKa = 3.52NIPATLTGSSVWKK154 pKa = 9.85NRR156 pKa = 11.84GVIADD161 pKa = 3.74TATSVKK167 pKa = 10.31APGRR171 pKa = 11.84ISCDD175 pKa = 3.25AVCSYY180 pKa = 11.01CITFISFCFFHH191 pKa = 7.8SSALFKK197 pKa = 10.99VPKK200 pKa = 9.56PLLNFEE206 pKa = 4.2TAVAYY211 pKa = 10.62SLVLQVEE218 pKa = 4.53LEE220 pKa = 4.08FPNIKK225 pKa = 9.15DD226 pKa = 3.71TLHH229 pKa = 6.46EE230 pKa = 4.86KK231 pKa = 10.12YY232 pKa = 10.72LKK234 pKa = 11.01NKK236 pKa = 9.43DD237 pKa = 3.87SKK239 pKa = 10.11WYY241 pKa = 8.35CTIDD245 pKa = 3.09IHH247 pKa = 8.41IGNLLKK253 pKa = 10.05RR254 pKa = 11.84TAKK257 pKa = 9.2QRR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84TPSEE265 pKa = 3.25ITQKK269 pKa = 9.29VRR271 pKa = 11.84RR272 pKa = 11.84MGFRR276 pKa = 11.84IGLYY280 pKa = 9.89DD281 pKa = 3.61LWGPTIVVEE290 pKa = 4.17LTGSSSKK297 pKa = 10.74SLQGFFSSEE306 pKa = 3.47RR307 pKa = 11.84LACHH311 pKa = 7.22PISQYY316 pKa = 10.74NPHH319 pKa = 6.29VGQLIWAHH327 pKa = 6.2DD328 pKa = 3.73VSITGCHH335 pKa = 5.71MIISEE340 pKa = 4.31LEE342 pKa = 4.02KK343 pKa = 10.93KK344 pKa = 9.98KK345 pKa = 10.84ALAMADD351 pKa = 3.78LTVSDD356 pKa = 4.43AVAINTTIKK365 pKa = 10.42EE366 pKa = 4.2LVPFRR371 pKa = 11.84LFRR374 pKa = 11.84KK375 pKa = 9.62
MM1 pKa = 7.55AAHH4 pKa = 7.22PNHH7 pKa = 6.99ANPSSSISLMHH18 pKa = 7.63DD19 pKa = 3.5DD20 pKa = 5.63PSIQTQLLAFPLISEE35 pKa = 4.31KK36 pKa = 10.54TEE38 pKa = 3.77TGTTKK43 pKa = 10.57LQPQVRR49 pKa = 11.84MQSFLSTDD57 pKa = 3.07SQKK60 pKa = 10.89YY61 pKa = 9.26HH62 pKa = 6.38LVFINTYY69 pKa = 10.34GFIAEE74 pKa = 5.21DD75 pKa = 4.69FNCSPTNGFVPALFQPKK92 pKa = 10.21SKK94 pKa = 10.29VLSSAMVTLGAVPADD109 pKa = 3.67TVLQDD114 pKa = 3.67LQKK117 pKa = 11.21DD118 pKa = 4.44LIAMRR123 pKa = 11.84FKK125 pKa = 10.78VRR127 pKa = 11.84KK128 pKa = 8.73SASAKK133 pKa = 10.0EE134 pKa = 4.36LILFSTDD141 pKa = 3.52NIPATLTGSSVWKK154 pKa = 9.85NRR156 pKa = 11.84GVIADD161 pKa = 3.74TATSVKK167 pKa = 10.31APGRR171 pKa = 11.84ISCDD175 pKa = 3.25AVCSYY180 pKa = 11.01CITFISFCFFHH191 pKa = 7.8SSALFKK197 pKa = 10.99VPKK200 pKa = 9.56PLLNFEE206 pKa = 4.2TAVAYY211 pKa = 10.62SLVLQVEE218 pKa = 4.53LEE220 pKa = 4.08FPNIKK225 pKa = 9.15DD226 pKa = 3.71TLHH229 pKa = 6.46EE230 pKa = 4.86KK231 pKa = 10.12YY232 pKa = 10.72LKK234 pKa = 11.01NKK236 pKa = 9.43DD237 pKa = 3.87SKK239 pKa = 10.11WYY241 pKa = 8.35CTIDD245 pKa = 3.09IHH247 pKa = 8.41IGNLLKK253 pKa = 10.05RR254 pKa = 11.84TAKK257 pKa = 9.2QRR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84TPSEE265 pKa = 3.25ITQKK269 pKa = 9.29VRR271 pKa = 11.84RR272 pKa = 11.84MGFRR276 pKa = 11.84IGLYY280 pKa = 9.89DD281 pKa = 3.61LWGPTIVVEE290 pKa = 4.17LTGSSSKK297 pKa = 10.74SLQGFFSSEE306 pKa = 3.47RR307 pKa = 11.84LACHH311 pKa = 7.22PISQYY316 pKa = 10.74NPHH319 pKa = 6.29VGQLIWAHH327 pKa = 6.2DD328 pKa = 3.73VSITGCHH335 pKa = 5.71MIISEE340 pKa = 4.31LEE342 pKa = 4.02KK343 pKa = 10.93KK344 pKa = 9.98KK345 pKa = 10.84ALAMADD351 pKa = 3.78LTVSDD356 pKa = 4.43AVAINTTIKK365 pKa = 10.42EE366 pKa = 4.2LVPFRR371 pKa = 11.84LFRR374 pKa = 11.84KK375 pKa = 9.62
Molecular weight: 41.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4535 |
375 |
2198 |
755.8 |
84.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.086 ± 0.626 | 2.426 ± 0.337 |
5.006 ± 0.36 | 4.3 ± 0.295 |
3.462 ± 0.557 | 4.939 ± 0.311 |
2.492 ± 0.262 | 8.093 ± 0.872 |
4.19 ± 0.525 | 11.51 ± 1.231 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.073 ± 0.367 | 5.05 ± 0.311 |
4.851 ± 0.585 | 4.564 ± 0.371 |
4.917 ± 0.351 | 9.46 ± 0.663 |
7.1 ± 0.492 | 5.204 ± 0.265 |
0.794 ± 0.169 | 3.44 ± 0.461 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
