
Treponema azotonutricium (strain ATCC BAA-888 / DSM 13862 / ZAS-9)
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Treponemataceae; Treponema; Treponema azotonutricium
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
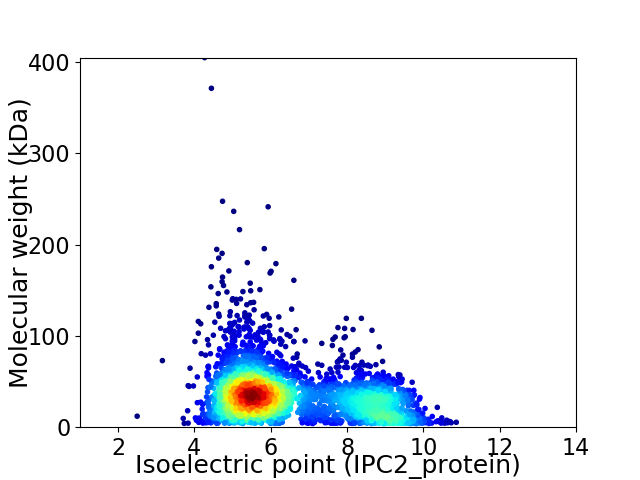
Virtual 2D-PAGE plot for 3457 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F5Y8N8|F5Y8N8_TREAZ Undecaprenyl-diphosphatase OS=Treponema azotonutricium (strain ATCC BAA-888 / DSM 13862 / ZAS-9) OX=545695 GN=uppP PE=3 SV=1
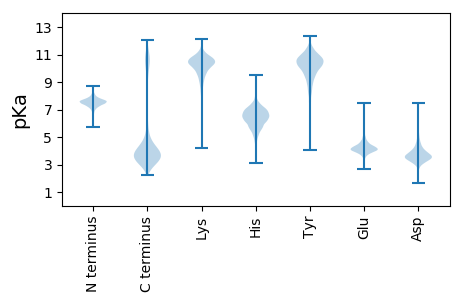
MM1 pKa = 7.17KK2 pKa = 10.21HH3 pKa = 6.67LIIALLLSALLFAACSGKK21 pKa = 10.23KK22 pKa = 8.98EE23 pKa = 3.74AARR26 pKa = 11.84NSDD29 pKa = 2.66ASEE32 pKa = 4.01AVLGDD37 pKa = 3.33VDD39 pKa = 4.26EE40 pKa = 5.58DD41 pKa = 4.03LVSAALFAIYY51 pKa = 9.44AQKK54 pKa = 9.3GTPEE58 pKa = 3.72WNALDD63 pKa = 3.55SLGVHH68 pKa = 7.02TIATDD73 pKa = 4.2FSTPGAGGAVRR84 pKa = 11.84VQLLQDD90 pKa = 4.11LSSFMLAAVTFNDD103 pKa = 3.88EE104 pKa = 5.49DD105 pKa = 4.0EE106 pKa = 5.21DD107 pKa = 4.92FYY109 pKa = 11.12PSYY112 pKa = 11.63VFFDD116 pKa = 3.68IPGKK120 pKa = 10.69AGDD123 pKa = 3.89VYY125 pKa = 11.07EE126 pKa = 4.56FDD128 pKa = 5.38AIFTEE133 pKa = 5.52GIPLGSITAVRR144 pKa = 11.84SDD146 pKa = 3.04NRR148 pKa = 11.84GNGDD152 pKa = 3.66PYY154 pKa = 8.32TTYY157 pKa = 11.26YY158 pKa = 10.1LQEE161 pKa = 4.31SGFDD165 pKa = 3.61GLPNGLVVYY174 pKa = 10.6NEE176 pKa = 4.29EE177 pKa = 4.35FNSQEE182 pKa = 4.45DD183 pKa = 3.61EE184 pKa = 4.23WEE186 pKa = 4.34EE187 pKa = 3.81PSAPPDD193 pKa = 3.0QDD195 pKa = 4.34AIYY198 pKa = 10.91DD199 pKa = 4.06EE200 pKa = 5.71LLDD203 pKa = 4.91NQDD206 pKa = 2.57AWFAGLKK213 pKa = 8.53ATSPFGSYY221 pKa = 10.39DD222 pKa = 3.53RR223 pKa = 11.84NIFSGGDD230 pKa = 3.29FYY232 pKa = 11.66LGVGITLDD240 pKa = 3.38IDD242 pKa = 3.84GDD244 pKa = 4.56GIFEE248 pKa = 4.42LWFIATNQDD257 pKa = 2.58RR258 pKa = 11.84SEE260 pKa = 4.19QYY262 pKa = 9.12TFLYY266 pKa = 9.73TLVEE270 pKa = 4.17GKK272 pKa = 9.07PLCLLSEE279 pKa = 4.27LTLTYY284 pKa = 10.98EE285 pKa = 4.41DD286 pKa = 5.38LSDD289 pKa = 3.82NLLGFIDD296 pKa = 3.9VVYY299 pKa = 10.55DD300 pKa = 3.73YY301 pKa = 9.03DD302 pKa = 3.84TLTQRR307 pKa = 11.84IVFVRR312 pKa = 11.84GNIRR316 pKa = 11.84GFPGWSRR323 pKa = 11.84RR324 pKa = 11.84DD325 pKa = 3.08VYY327 pKa = 11.26YY328 pKa = 10.39RR329 pKa = 11.84YY330 pKa = 10.52SRR332 pKa = 11.84GKK334 pKa = 9.7LDD336 pKa = 4.8KK337 pKa = 10.87IADD340 pKa = 4.17LLSLNGSRR348 pKa = 11.84NEE350 pKa = 3.8LEE352 pKa = 3.83AAEE355 pKa = 4.54FLTGPEE361 pKa = 4.5FYY363 pKa = 10.13TGNGDD368 pKa = 4.13DD369 pKa = 5.41DD370 pKa = 3.59ITVFFINGKK379 pKa = 9.53AATADD384 pKa = 4.11DD385 pKa = 3.79YY386 pKa = 11.89TLIVDD391 pKa = 4.23SYY393 pKa = 9.98VGSVLSRR400 pKa = 11.84YY401 pKa = 8.16YY402 pKa = 11.46ANN404 pKa = 3.96
MM1 pKa = 7.17KK2 pKa = 10.21HH3 pKa = 6.67LIIALLLSALLFAACSGKK21 pKa = 10.23KK22 pKa = 8.98EE23 pKa = 3.74AARR26 pKa = 11.84NSDD29 pKa = 2.66ASEE32 pKa = 4.01AVLGDD37 pKa = 3.33VDD39 pKa = 4.26EE40 pKa = 5.58DD41 pKa = 4.03LVSAALFAIYY51 pKa = 9.44AQKK54 pKa = 9.3GTPEE58 pKa = 3.72WNALDD63 pKa = 3.55SLGVHH68 pKa = 7.02TIATDD73 pKa = 4.2FSTPGAGGAVRR84 pKa = 11.84VQLLQDD90 pKa = 4.11LSSFMLAAVTFNDD103 pKa = 3.88EE104 pKa = 5.49DD105 pKa = 4.0EE106 pKa = 5.21DD107 pKa = 4.92FYY109 pKa = 11.12PSYY112 pKa = 11.63VFFDD116 pKa = 3.68IPGKK120 pKa = 10.69AGDD123 pKa = 3.89VYY125 pKa = 11.07EE126 pKa = 4.56FDD128 pKa = 5.38AIFTEE133 pKa = 5.52GIPLGSITAVRR144 pKa = 11.84SDD146 pKa = 3.04NRR148 pKa = 11.84GNGDD152 pKa = 3.66PYY154 pKa = 8.32TTYY157 pKa = 11.26YY158 pKa = 10.1LQEE161 pKa = 4.31SGFDD165 pKa = 3.61GLPNGLVVYY174 pKa = 10.6NEE176 pKa = 4.29EE177 pKa = 4.35FNSQEE182 pKa = 4.45DD183 pKa = 3.61EE184 pKa = 4.23WEE186 pKa = 4.34EE187 pKa = 3.81PSAPPDD193 pKa = 3.0QDD195 pKa = 4.34AIYY198 pKa = 10.91DD199 pKa = 4.06EE200 pKa = 5.71LLDD203 pKa = 4.91NQDD206 pKa = 2.57AWFAGLKK213 pKa = 8.53ATSPFGSYY221 pKa = 10.39DD222 pKa = 3.53RR223 pKa = 11.84NIFSGGDD230 pKa = 3.29FYY232 pKa = 11.66LGVGITLDD240 pKa = 3.38IDD242 pKa = 3.84GDD244 pKa = 4.56GIFEE248 pKa = 4.42LWFIATNQDD257 pKa = 2.58RR258 pKa = 11.84SEE260 pKa = 4.19QYY262 pKa = 9.12TFLYY266 pKa = 9.73TLVEE270 pKa = 4.17GKK272 pKa = 9.07PLCLLSEE279 pKa = 4.27LTLTYY284 pKa = 10.98EE285 pKa = 4.41DD286 pKa = 5.38LSDD289 pKa = 3.82NLLGFIDD296 pKa = 3.9VVYY299 pKa = 10.55DD300 pKa = 3.73YY301 pKa = 9.03DD302 pKa = 3.84TLTQRR307 pKa = 11.84IVFVRR312 pKa = 11.84GNIRR316 pKa = 11.84GFPGWSRR323 pKa = 11.84RR324 pKa = 11.84DD325 pKa = 3.08VYY327 pKa = 11.26YY328 pKa = 10.39RR329 pKa = 11.84YY330 pKa = 10.52SRR332 pKa = 11.84GKK334 pKa = 9.7LDD336 pKa = 4.8KK337 pKa = 10.87IADD340 pKa = 4.17LLSLNGSRR348 pKa = 11.84NEE350 pKa = 3.8LEE352 pKa = 3.83AAEE355 pKa = 4.54FLTGPEE361 pKa = 4.5FYY363 pKa = 10.13TGNGDD368 pKa = 4.13DD369 pKa = 5.41DD370 pKa = 3.59ITVFFINGKK379 pKa = 9.53AATADD384 pKa = 4.11DD385 pKa = 3.79YY386 pKa = 11.89TLIVDD391 pKa = 4.23SYY393 pKa = 9.98VGSVLSRR400 pKa = 11.84YY401 pKa = 8.16YY402 pKa = 11.46ANN404 pKa = 3.96
Molecular weight: 44.69 kDa
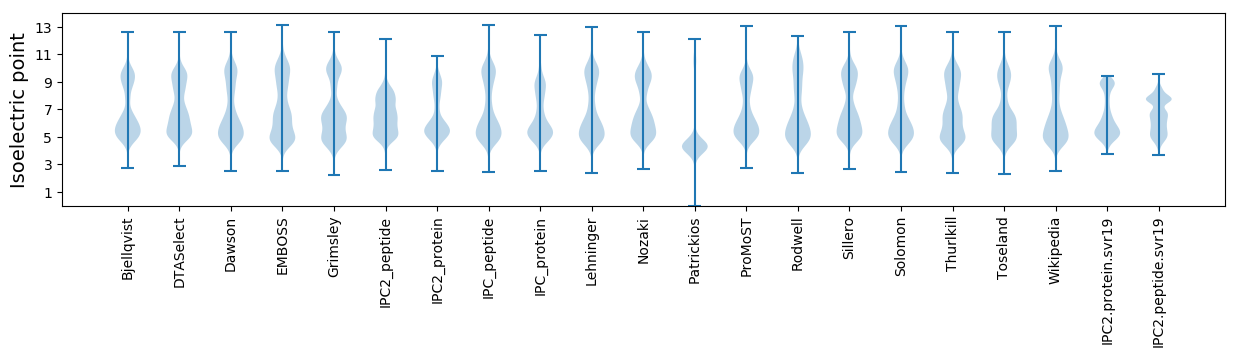
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F5YBK1|F5YBK1_TREAZ Putative TPR domain protein OS=Treponema azotonutricium (strain ATCC BAA-888 / DSM 13862 / ZAS-9) OX=545695 GN=TREAZ_0576 PE=4 SV=1
MM1 pKa = 7.52LGSSMGRR8 pKa = 11.84PSLFSWGRR16 pKa = 11.84MAFRR20 pKa = 11.84LLTNPAGGKK29 pKa = 9.0SGKK32 pKa = 9.69GGKK35 pKa = 8.48VWMRR39 pKa = 11.84QIMRR43 pKa = 11.84LMWTVV48 pKa = 2.74
MM1 pKa = 7.52LGSSMGRR8 pKa = 11.84PSLFSWGRR16 pKa = 11.84MAFRR20 pKa = 11.84LLTNPAGGKK29 pKa = 9.0SGKK32 pKa = 9.69GGKK35 pKa = 8.48VWMRR39 pKa = 11.84QIMRR43 pKa = 11.84LMWTVV48 pKa = 2.74
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1156691 |
37 |
4075 |
334.6 |
37.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.075 ± 0.053 | 1.044 ± 0.017 |
5.421 ± 0.03 | 6.593 ± 0.043 |
4.624 ± 0.035 | 7.78 ± 0.048 |
1.587 ± 0.016 | 7.234 ± 0.039 |
6.09 ± 0.037 | 9.644 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.383 ± 0.021 | 4.146 ± 0.037 |
4.359 ± 0.032 | 2.817 ± 0.023 |
5.003 ± 0.036 | 6.472 ± 0.041 |
5.032 ± 0.046 | 6.103 ± 0.034 |
1.147 ± 0.014 | 3.444 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
