
Beihai sobemo-like virus 17
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
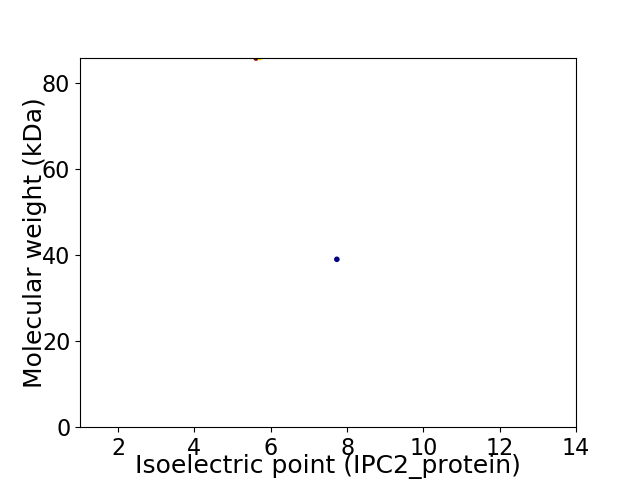
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEE0|A0A1L3KEE0_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 17 OX=1922688 PE=4 SV=1
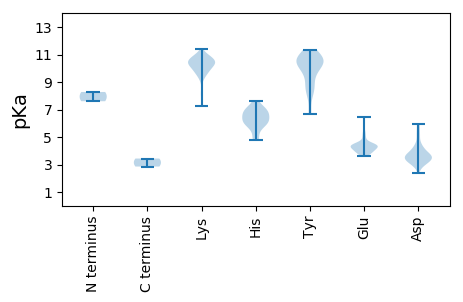
MM1 pKa = 7.6ISYY4 pKa = 8.49EE5 pKa = 4.07QLTGYY10 pKa = 10.4YY11 pKa = 8.67YY12 pKa = 10.75VASAVCSRR20 pKa = 11.84VLEE23 pKa = 4.41AFLEE27 pKa = 4.52AMLNWLLSPYY37 pKa = 10.17GRR39 pKa = 11.84GVTFFLILQLAVFSAVLAYY58 pKa = 10.6AFTYY62 pKa = 10.15YY63 pKa = 10.38AAKK66 pKa = 10.24RR67 pKa = 11.84VWTLVKK73 pKa = 10.09YY74 pKa = 10.3VVFIFLAPSYY84 pKa = 10.01YY85 pKa = 10.53VYY87 pKa = 10.85EE88 pKa = 4.35RR89 pKa = 11.84YY90 pKa = 9.78IKK92 pKa = 10.32GFEE95 pKa = 4.45LIYY98 pKa = 10.24PRR100 pKa = 11.84NLFVQEE106 pKa = 4.46SINYY110 pKa = 6.71LQRR113 pKa = 11.84VKK115 pKa = 10.79ALPQCQVILCTRR127 pKa = 11.84SDD129 pKa = 3.1EE130 pKa = 4.48SLFRR134 pKa = 11.84KK135 pKa = 9.77SYY137 pKa = 10.54RR138 pKa = 11.84VIGQAFFIGINQGGPDD154 pKa = 3.69TLMTAAHH161 pKa = 6.26NLVGQDD167 pKa = 2.86KK168 pKa = 10.33EE169 pKa = 4.4YY170 pKa = 10.84FITRR174 pKa = 11.84DD175 pKa = 3.2MKK177 pKa = 10.38TFHH180 pKa = 6.97PVLSNEE186 pKa = 4.06QDD188 pKa = 3.21SGSPVIDD195 pKa = 3.45VVLLKK200 pKa = 10.68VPQGTGTKK208 pKa = 9.92LRR210 pKa = 11.84AAPASTQCWCPNSSAVEE227 pKa = 4.35VYY229 pKa = 10.61AGGMAEE235 pKa = 4.0GGEE238 pKa = 4.32PVKK241 pKa = 11.0SVGQVTSIEE250 pKa = 4.22GPNGIFTLAHH260 pKa = 5.94SASTDD265 pKa = 3.19GGFSGAPLFQGNKK278 pKa = 7.27VVGMHH283 pKa = 7.07LASLQNQVNLGVWLWPILGEE303 pKa = 4.28VKK305 pKa = 10.26EE306 pKa = 4.15VTPRR310 pKa = 11.84GFGDD314 pKa = 4.33FSQEE318 pKa = 3.74SGEE321 pKa = 4.11FSLEE325 pKa = 3.78HH326 pKa = 7.64SEE328 pKa = 6.42DD329 pKa = 3.04GFAWMKK335 pKa = 10.65DD336 pKa = 3.46EE337 pKa = 5.06PPFEE341 pKa = 4.25LSVRR345 pKa = 11.84GDD347 pKa = 3.1RR348 pKa = 11.84AYY350 pKa = 11.34VRR352 pKa = 11.84TSDD355 pKa = 3.27GRR357 pKa = 11.84NYY359 pKa = 10.61NYY361 pKa = 9.44EE362 pKa = 4.04ATDD365 pKa = 2.89HH366 pKa = 6.6HH367 pKa = 7.08LRR369 pKa = 11.84LLRR372 pKa = 11.84SKK374 pKa = 10.67YY375 pKa = 8.56SQFRR379 pKa = 11.84KK380 pKa = 10.29SYY382 pKa = 11.26DD383 pKa = 3.25DD384 pKa = 5.33FIDD387 pKa = 3.76YY388 pKa = 10.8DD389 pKa = 3.74QVEE392 pKa = 4.33RR393 pKa = 11.84DD394 pKa = 4.07SNYY397 pKa = 8.4HH398 pKa = 4.81TAVGKK403 pKa = 10.09AWEE406 pKa = 4.08VDD408 pKa = 3.4EE409 pKa = 5.36PDD411 pKa = 3.64FDD413 pKa = 5.98EE414 pKa = 5.28VIQWEE419 pKa = 4.41NAKK422 pKa = 10.56KK423 pKa = 9.74MEE425 pKa = 4.52NEE427 pKa = 5.49SFLQWIKK434 pKa = 9.96QQIDD438 pKa = 3.56PKK440 pKa = 10.41NLVWEE445 pKa = 4.51SKK447 pKa = 9.38PRR449 pKa = 11.84PMEE452 pKa = 4.09NLEE455 pKa = 4.11PTHH458 pKa = 6.32VLTGLRR464 pKa = 11.84AFSRR468 pKa = 11.84ILHH471 pKa = 6.69DD472 pKa = 4.3ANEE475 pKa = 4.37AKK477 pKa = 10.59VAMEE481 pKa = 3.77MMCRR485 pKa = 11.84TGNLNLARR493 pKa = 11.84RR494 pKa = 11.84DD495 pKa = 3.63KK496 pKa = 10.84EE497 pKa = 4.53ASFQQAADD505 pKa = 3.56ASLLADD511 pKa = 3.84TMRR514 pKa = 11.84VISTKK519 pKa = 10.4LAEE522 pKa = 4.41ASDD525 pKa = 4.25LFEE528 pKa = 4.46QVVQKK533 pKa = 10.65SDD535 pKa = 2.97KK536 pKa = 10.96DD537 pKa = 3.63AAQSTNWANYY547 pKa = 6.73RR548 pKa = 11.84HH549 pKa = 6.23KK550 pKa = 10.3HH551 pKa = 5.84RR552 pKa = 11.84DD553 pKa = 3.63DD554 pKa = 3.73MPPPQAQTRR563 pKa = 11.84AHH565 pKa = 5.91VDD567 pKa = 2.41RR568 pKa = 11.84WLRR571 pKa = 11.84NIEE574 pKa = 3.94TQLIVLRR581 pKa = 11.84AVPKK585 pKa = 10.16RR586 pKa = 11.84QTPTSISQTAEE597 pKa = 3.68PQVVQEE603 pKa = 4.41RR604 pKa = 11.84LSDD607 pKa = 3.63LSDD610 pKa = 3.2MFEE613 pKa = 4.18TFGLNFEE620 pKa = 4.35EE621 pKa = 5.04TEE623 pKa = 4.28DD624 pKa = 3.98GQDD627 pKa = 3.42FRR629 pKa = 11.84LGAPQKK635 pKa = 10.94EE636 pKa = 4.61DD637 pKa = 3.49GAPDD641 pKa = 3.5NQKK644 pKa = 10.9AEE646 pKa = 4.47LSSQPSLCSEE656 pKa = 4.21EE657 pKa = 4.26QSSEE661 pKa = 4.14TIQKK665 pKa = 9.45KK666 pKa = 10.37KK667 pKa = 9.78KK668 pKa = 10.51VSWAPNLEE676 pKa = 4.51TPSEE680 pKa = 4.34TRR682 pKa = 11.84NSRR685 pKa = 11.84HH686 pKa = 5.34QNTSRR691 pKa = 11.84KK692 pKa = 9.23PSQEE696 pKa = 3.94KK697 pKa = 11.02NNMGGPRR704 pKa = 11.84KK705 pKa = 9.23PRR707 pKa = 11.84NPRR710 pKa = 11.84RR711 pKa = 11.84FEE713 pKa = 3.71KK714 pKa = 10.52RR715 pKa = 11.84SNFTQEE721 pKa = 4.62SMMPPDD727 pKa = 3.79NGVEE731 pKa = 3.98EE732 pKa = 4.31LLKK735 pKa = 10.35KK736 pKa = 10.95VSMAALKK743 pKa = 10.43PFFEE747 pKa = 4.35ATLRR751 pKa = 11.84QLDD754 pKa = 3.66NSQQ757 pKa = 2.85
MM1 pKa = 7.6ISYY4 pKa = 8.49EE5 pKa = 4.07QLTGYY10 pKa = 10.4YY11 pKa = 8.67YY12 pKa = 10.75VASAVCSRR20 pKa = 11.84VLEE23 pKa = 4.41AFLEE27 pKa = 4.52AMLNWLLSPYY37 pKa = 10.17GRR39 pKa = 11.84GVTFFLILQLAVFSAVLAYY58 pKa = 10.6AFTYY62 pKa = 10.15YY63 pKa = 10.38AAKK66 pKa = 10.24RR67 pKa = 11.84VWTLVKK73 pKa = 10.09YY74 pKa = 10.3VVFIFLAPSYY84 pKa = 10.01YY85 pKa = 10.53VYY87 pKa = 10.85EE88 pKa = 4.35RR89 pKa = 11.84YY90 pKa = 9.78IKK92 pKa = 10.32GFEE95 pKa = 4.45LIYY98 pKa = 10.24PRR100 pKa = 11.84NLFVQEE106 pKa = 4.46SINYY110 pKa = 6.71LQRR113 pKa = 11.84VKK115 pKa = 10.79ALPQCQVILCTRR127 pKa = 11.84SDD129 pKa = 3.1EE130 pKa = 4.48SLFRR134 pKa = 11.84KK135 pKa = 9.77SYY137 pKa = 10.54RR138 pKa = 11.84VIGQAFFIGINQGGPDD154 pKa = 3.69TLMTAAHH161 pKa = 6.26NLVGQDD167 pKa = 2.86KK168 pKa = 10.33EE169 pKa = 4.4YY170 pKa = 10.84FITRR174 pKa = 11.84DD175 pKa = 3.2MKK177 pKa = 10.38TFHH180 pKa = 6.97PVLSNEE186 pKa = 4.06QDD188 pKa = 3.21SGSPVIDD195 pKa = 3.45VVLLKK200 pKa = 10.68VPQGTGTKK208 pKa = 9.92LRR210 pKa = 11.84AAPASTQCWCPNSSAVEE227 pKa = 4.35VYY229 pKa = 10.61AGGMAEE235 pKa = 4.0GGEE238 pKa = 4.32PVKK241 pKa = 11.0SVGQVTSIEE250 pKa = 4.22GPNGIFTLAHH260 pKa = 5.94SASTDD265 pKa = 3.19GGFSGAPLFQGNKK278 pKa = 7.27VVGMHH283 pKa = 7.07LASLQNQVNLGVWLWPILGEE303 pKa = 4.28VKK305 pKa = 10.26EE306 pKa = 4.15VTPRR310 pKa = 11.84GFGDD314 pKa = 4.33FSQEE318 pKa = 3.74SGEE321 pKa = 4.11FSLEE325 pKa = 3.78HH326 pKa = 7.64SEE328 pKa = 6.42DD329 pKa = 3.04GFAWMKK335 pKa = 10.65DD336 pKa = 3.46EE337 pKa = 5.06PPFEE341 pKa = 4.25LSVRR345 pKa = 11.84GDD347 pKa = 3.1RR348 pKa = 11.84AYY350 pKa = 11.34VRR352 pKa = 11.84TSDD355 pKa = 3.27GRR357 pKa = 11.84NYY359 pKa = 10.61NYY361 pKa = 9.44EE362 pKa = 4.04ATDD365 pKa = 2.89HH366 pKa = 6.6HH367 pKa = 7.08LRR369 pKa = 11.84LLRR372 pKa = 11.84SKK374 pKa = 10.67YY375 pKa = 8.56SQFRR379 pKa = 11.84KK380 pKa = 10.29SYY382 pKa = 11.26DD383 pKa = 3.25DD384 pKa = 5.33FIDD387 pKa = 3.76YY388 pKa = 10.8DD389 pKa = 3.74QVEE392 pKa = 4.33RR393 pKa = 11.84DD394 pKa = 4.07SNYY397 pKa = 8.4HH398 pKa = 4.81TAVGKK403 pKa = 10.09AWEE406 pKa = 4.08VDD408 pKa = 3.4EE409 pKa = 5.36PDD411 pKa = 3.64FDD413 pKa = 5.98EE414 pKa = 5.28VIQWEE419 pKa = 4.41NAKK422 pKa = 10.56KK423 pKa = 9.74MEE425 pKa = 4.52NEE427 pKa = 5.49SFLQWIKK434 pKa = 9.96QQIDD438 pKa = 3.56PKK440 pKa = 10.41NLVWEE445 pKa = 4.51SKK447 pKa = 9.38PRR449 pKa = 11.84PMEE452 pKa = 4.09NLEE455 pKa = 4.11PTHH458 pKa = 6.32VLTGLRR464 pKa = 11.84AFSRR468 pKa = 11.84ILHH471 pKa = 6.69DD472 pKa = 4.3ANEE475 pKa = 4.37AKK477 pKa = 10.59VAMEE481 pKa = 3.77MMCRR485 pKa = 11.84TGNLNLARR493 pKa = 11.84RR494 pKa = 11.84DD495 pKa = 3.63KK496 pKa = 10.84EE497 pKa = 4.53ASFQQAADD505 pKa = 3.56ASLLADD511 pKa = 3.84TMRR514 pKa = 11.84VISTKK519 pKa = 10.4LAEE522 pKa = 4.41ASDD525 pKa = 4.25LFEE528 pKa = 4.46QVVQKK533 pKa = 10.65SDD535 pKa = 2.97KK536 pKa = 10.96DD537 pKa = 3.63AAQSTNWANYY547 pKa = 6.73RR548 pKa = 11.84HH549 pKa = 6.23KK550 pKa = 10.3HH551 pKa = 5.84RR552 pKa = 11.84DD553 pKa = 3.63DD554 pKa = 3.73MPPPQAQTRR563 pKa = 11.84AHH565 pKa = 5.91VDD567 pKa = 2.41RR568 pKa = 11.84WLRR571 pKa = 11.84NIEE574 pKa = 3.94TQLIVLRR581 pKa = 11.84AVPKK585 pKa = 10.16RR586 pKa = 11.84QTPTSISQTAEE597 pKa = 3.68PQVVQEE603 pKa = 4.41RR604 pKa = 11.84LSDD607 pKa = 3.63LSDD610 pKa = 3.2MFEE613 pKa = 4.18TFGLNFEE620 pKa = 4.35EE621 pKa = 5.04TEE623 pKa = 4.28DD624 pKa = 3.98GQDD627 pKa = 3.42FRR629 pKa = 11.84LGAPQKK635 pKa = 10.94EE636 pKa = 4.61DD637 pKa = 3.49GAPDD641 pKa = 3.5NQKK644 pKa = 10.9AEE646 pKa = 4.47LSSQPSLCSEE656 pKa = 4.21EE657 pKa = 4.26QSSEE661 pKa = 4.14TIQKK665 pKa = 9.45KK666 pKa = 10.37KK667 pKa = 9.78KK668 pKa = 10.51VSWAPNLEE676 pKa = 4.51TPSEE680 pKa = 4.34TRR682 pKa = 11.84NSRR685 pKa = 11.84HH686 pKa = 5.34QNTSRR691 pKa = 11.84KK692 pKa = 9.23PSQEE696 pKa = 3.94KK697 pKa = 11.02NNMGGPRR704 pKa = 11.84KK705 pKa = 9.23PRR707 pKa = 11.84NPRR710 pKa = 11.84RR711 pKa = 11.84FEE713 pKa = 3.71KK714 pKa = 10.52RR715 pKa = 11.84SNFTQEE721 pKa = 4.62SMMPPDD727 pKa = 3.79NGVEE731 pKa = 3.98EE732 pKa = 4.31LLKK735 pKa = 10.35KK736 pKa = 10.95VSMAALKK743 pKa = 10.43PFFEE747 pKa = 4.35ATLRR751 pKa = 11.84QLDD754 pKa = 3.66NSQQ757 pKa = 2.85
Molecular weight: 85.91 kDa
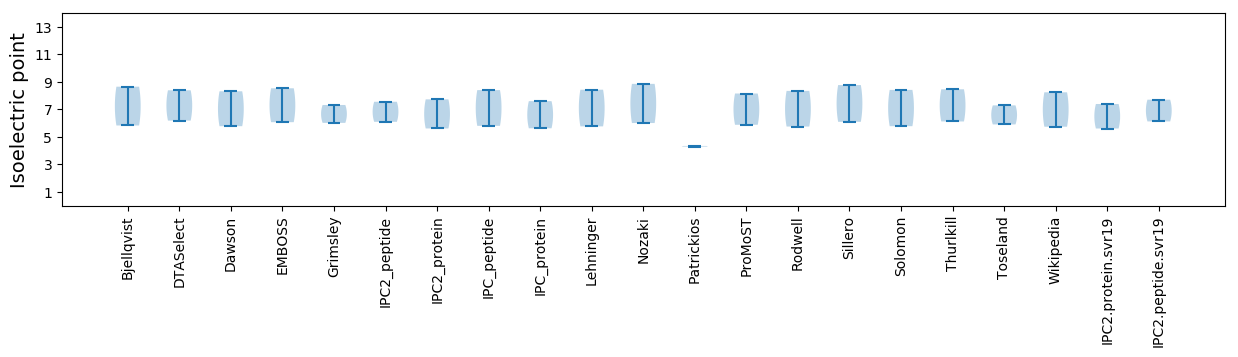
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEE0|A0A1L3KEE0_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 17 OX=1922688 PE=4 SV=1
MM1 pKa = 8.28RR2 pKa = 11.84DD3 pKa = 3.82GTPMLDD9 pKa = 3.63PKK11 pKa = 10.72SCPGNPWVSLGSTNEE26 pKa = 3.79VVFGYY31 pKa = 10.67LPGFGYY37 pKa = 10.55QGDD40 pKa = 4.11RR41 pKa = 11.84VNDD44 pKa = 4.16LYY46 pKa = 11.26HH47 pKa = 7.09AVLNRR52 pKa = 11.84FKK54 pKa = 10.83QLEE57 pKa = 4.3TAPAMDD63 pKa = 5.76PIFPFIKK70 pKa = 10.26QEE72 pKa = 3.87PHH74 pKa = 6.65KK75 pKa = 9.78KK76 pKa = 8.34TKK78 pKa = 10.66LEE80 pKa = 3.96EE81 pKa = 3.99GRR83 pKa = 11.84VRR85 pKa = 11.84IISGVSATDD94 pKa = 3.37QIVADD99 pKa = 4.34LLFMPEE105 pKa = 4.83LQSQMQNTLYY115 pKa = 10.93NGIAIGWAPNVPSGIVAFNEE135 pKa = 3.99VTEE138 pKa = 4.95GFDD141 pKa = 3.13TCADD145 pKa = 3.41KK146 pKa = 11.38SSWDD150 pKa = 3.34WTVVLWMVNFACQFFVSRR168 pKa = 11.84ATGCHH173 pKa = 4.83EE174 pKa = 3.75VLKK177 pKa = 10.72NHH179 pKa = 6.72ILAMLGPKK187 pKa = 9.6KK188 pKa = 10.45FKK190 pKa = 10.59FRR192 pKa = 11.84GGVFRR197 pKa = 11.84TEE199 pKa = 4.32HH200 pKa = 6.55PGVMPSGWKK209 pKa = 8.0VTILLNSLWQKK220 pKa = 10.38ACHH223 pKa = 6.01IMCGSPGKK231 pKa = 9.73ILSMGDD237 pKa = 3.28DD238 pKa = 4.56TIQEE242 pKa = 4.4PVQDD246 pKa = 4.19SYY248 pKa = 10.39WDD250 pKa = 3.43KK251 pKa = 10.48MAKK254 pKa = 9.91LGPKK258 pKa = 9.46IKK260 pKa = 10.15EE261 pKa = 3.63IRR263 pKa = 11.84KK264 pKa = 9.88DD265 pKa = 3.6GVKK268 pKa = 9.83EE269 pKa = 3.92FCGVTIDD276 pKa = 3.46KK277 pKa = 10.78NGYY280 pKa = 8.0YY281 pKa = 9.55PVYY284 pKa = 8.35TAKK287 pKa = 10.4HH288 pKa = 6.08SYY290 pKa = 9.46VLHH293 pKa = 5.99HH294 pKa = 7.0LKK296 pKa = 10.22PAVAIDD302 pKa = 3.59TLRR305 pKa = 11.84SYY307 pKa = 8.92QTLYY311 pKa = 10.79AYY313 pKa = 10.24CPKK316 pKa = 10.05QLSAIRR322 pKa = 11.84RR323 pKa = 11.84WLRR326 pKa = 11.84EE327 pKa = 3.78LGRR330 pKa = 11.84ADD332 pKa = 5.86AILPDD337 pKa = 4.86DD338 pKa = 5.17DD339 pKa = 5.2LRR341 pKa = 11.84KK342 pKa = 9.7PPLGLGG348 pKa = 3.44
MM1 pKa = 8.28RR2 pKa = 11.84DD3 pKa = 3.82GTPMLDD9 pKa = 3.63PKK11 pKa = 10.72SCPGNPWVSLGSTNEE26 pKa = 3.79VVFGYY31 pKa = 10.67LPGFGYY37 pKa = 10.55QGDD40 pKa = 4.11RR41 pKa = 11.84VNDD44 pKa = 4.16LYY46 pKa = 11.26HH47 pKa = 7.09AVLNRR52 pKa = 11.84FKK54 pKa = 10.83QLEE57 pKa = 4.3TAPAMDD63 pKa = 5.76PIFPFIKK70 pKa = 10.26QEE72 pKa = 3.87PHH74 pKa = 6.65KK75 pKa = 9.78KK76 pKa = 8.34TKK78 pKa = 10.66LEE80 pKa = 3.96EE81 pKa = 3.99GRR83 pKa = 11.84VRR85 pKa = 11.84IISGVSATDD94 pKa = 3.37QIVADD99 pKa = 4.34LLFMPEE105 pKa = 4.83LQSQMQNTLYY115 pKa = 10.93NGIAIGWAPNVPSGIVAFNEE135 pKa = 3.99VTEE138 pKa = 4.95GFDD141 pKa = 3.13TCADD145 pKa = 3.41KK146 pKa = 11.38SSWDD150 pKa = 3.34WTVVLWMVNFACQFFVSRR168 pKa = 11.84ATGCHH173 pKa = 4.83EE174 pKa = 3.75VLKK177 pKa = 10.72NHH179 pKa = 6.72ILAMLGPKK187 pKa = 9.6KK188 pKa = 10.45FKK190 pKa = 10.59FRR192 pKa = 11.84GGVFRR197 pKa = 11.84TEE199 pKa = 4.32HH200 pKa = 6.55PGVMPSGWKK209 pKa = 8.0VTILLNSLWQKK220 pKa = 10.38ACHH223 pKa = 6.01IMCGSPGKK231 pKa = 9.73ILSMGDD237 pKa = 3.28DD238 pKa = 4.56TIQEE242 pKa = 4.4PVQDD246 pKa = 4.19SYY248 pKa = 10.39WDD250 pKa = 3.43KK251 pKa = 10.48MAKK254 pKa = 9.91LGPKK258 pKa = 9.46IKK260 pKa = 10.15EE261 pKa = 3.63IRR263 pKa = 11.84KK264 pKa = 9.88DD265 pKa = 3.6GVKK268 pKa = 9.83EE269 pKa = 3.92FCGVTIDD276 pKa = 3.46KK277 pKa = 10.78NGYY280 pKa = 8.0YY281 pKa = 9.55PVYY284 pKa = 8.35TAKK287 pKa = 10.4HH288 pKa = 6.08SYY290 pKa = 9.46VLHH293 pKa = 5.99HH294 pKa = 7.0LKK296 pKa = 10.22PAVAIDD302 pKa = 3.59TLRR305 pKa = 11.84SYY307 pKa = 8.92QTLYY311 pKa = 10.79AYY313 pKa = 10.24CPKK316 pKa = 10.05QLSAIRR322 pKa = 11.84RR323 pKa = 11.84WLRR326 pKa = 11.84EE327 pKa = 3.78LGRR330 pKa = 11.84ADD332 pKa = 5.86AILPDD337 pKa = 4.86DD338 pKa = 5.17DD339 pKa = 5.2LRR341 pKa = 11.84KK342 pKa = 9.7PPLGLGG348 pKa = 3.44
Molecular weight: 39.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1105 |
348 |
757 |
552.5 |
62.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.059 ± 0.609 | 1.357 ± 0.56 |
5.52 ± 0.306 | 6.425 ± 1.428 |
4.706 ± 0.235 | 6.425 ± 1.134 |
2.081 ± 0.3 | 3.982 ± 0.879 |
5.973 ± 0.72 | 8.507 ± 0.239 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.715 ± 0.265 | 4.253 ± 0.479 |
5.792 ± 0.657 | 5.339 ± 0.953 |
5.43 ± 0.666 | 7.059 ± 1.121 |
4.977 ± 0.055 | 6.968 ± 0.128 |
1.991 ± 0.354 | 3.439 ± 0.006 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
